CopciAB_435561
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_435561 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 435561 |
| Uniprot id | Functional description | Aryl-alcohol oxidase | |
| Location | scaffold_11:814222..816620 | Strand | - |
| Gene length (nt) | 2399 | Transcript length (nt) | 1944 |
| CDS length (nt) | 1791 | Protein length (aa) | 596 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_217506 | 48.4 | 7.941E-176 | 559 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1109867 | 48.7 | 5.251E-173 | 551 |
| Pleurotus ostreatus PC9 | PleosPC9_1_100509 | 50 | 2.512E-172 | 549 |
| Pleurotus eryngii ATCC 90797 | Pleery1_871357 | 48.9 | 1.593E-170 | 544 |
| Flammulina velutipes | Flave_chr07AA01044 | 45.5 | 2.685E-162 | 520 |
| Agrocybe aegerita | Agrae_CAA7257390 | 46.5 | 4.099E-158 | 508 |
| Lentinula edodes B17 | Lened_B_1_1_8257 | 45.4 | 8.556E-155 | 498 |
| Schizophyllum commune H4-8 | Schco3_2642607 | 45.2 | 3.136E-152 | 491 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_435561.T0 |
| Description | Aryl-alcohol oxidase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | 37 | 351 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | 450 | 588 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 23 | 0.9883 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal |
| IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal |
| IPR036188 | FAD/NAD(P)-binding domain superfamily |
| IPR012132 | Glucose-methanol-choline oxidoreductase |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF |
| GO:0050660 | flavin adenine dinucleotide binding | MF |
KEGG
| KEGG Orthology |
|---|
| K00108 |
EggNOG
| COG category | Description |
|---|---|
| E | Aryl-alcohol oxidase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA3 | AA3_2 |
Transcription factor
| Group |
|---|
| No records |
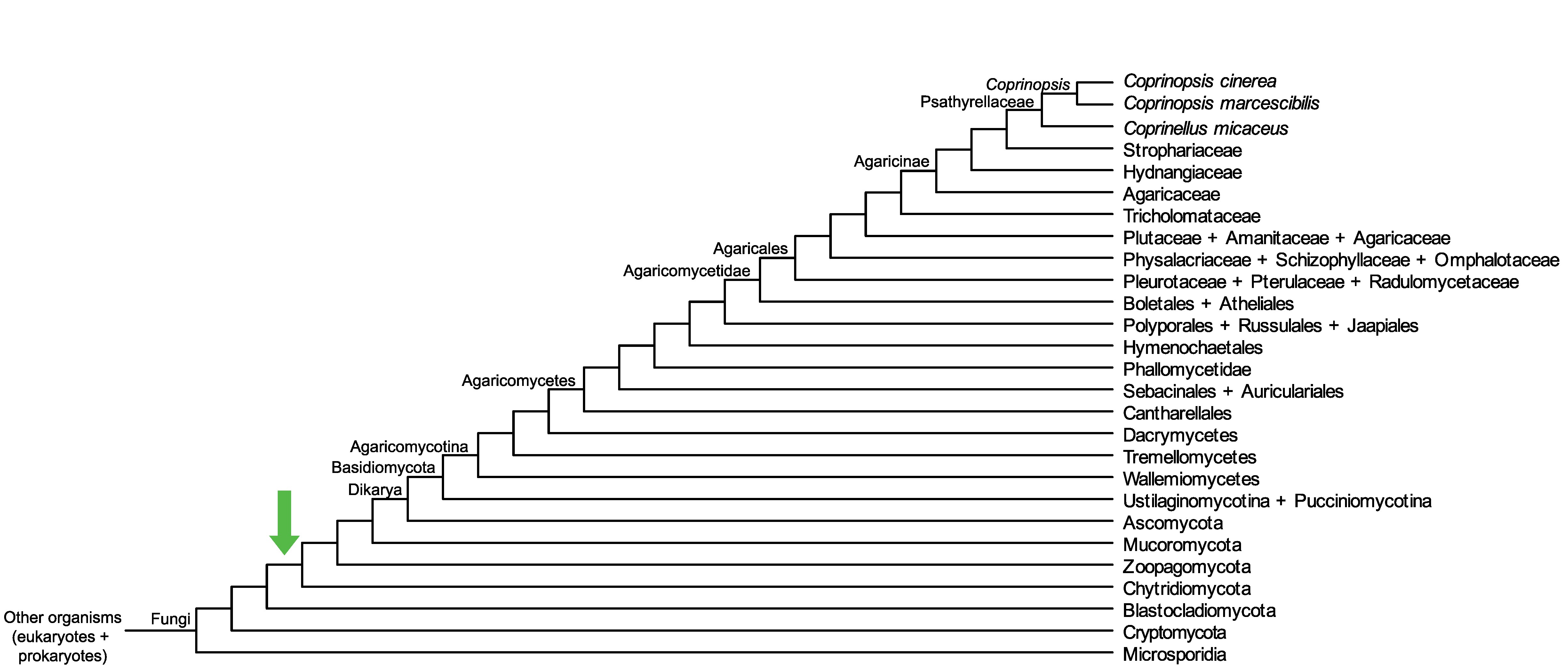
Conservation of CopciAB_435561 across fungi.
Arrow shows the origin of gene family containing CopciAB_435561.
Protein
| Sequence id | CopciAB_435561.T0 |
|---|---|
| Sequence |
>CopciAB_435561.T0 MGTRSLLPSLLVLASFLFHPSLAVIRDDFTDLPRLAYDFVIVGGGTAGSVLASRLTEDPDFNVLLIEAGPSHEGV LESQVPGLTFALQQTQYDWNFNTVPQIGLNNRTDRLPRGRMLGGSSSINGLFYTRGSSDDYDRWAAITGDSGWSW SNILPYLLKSEKWNPPTDGHDTTGKYDPSYHSTTGKVGVSLSGVPQASDSKVFEASQELGGEFAYDVDMNDGTPL GFGWLQSTINNGERSSAATGYLDATVLARPNLHVLVNNRVVKLAERKQPGSNTPAFQTVHFQTGGSPSGRLHRVT AKKEVILAAGAYGSPQLLMLSGIGDSTELQNIGIEPKVKLPSVGKNLTEHVQFSMMFSLGINDTVDPFLNQTFLM ESMQEWFDHRTGYMTNLGINSVIWHRLPEDWPHWSQFEDPTGGINSPHIELVVMGGGIYPTPGPNLIVSPVLASP RSRGFMTLNSTSPYASPLIDPAMLTHPFDILALRESVRSVLKFVGASTWDEYNLTLLPPFAGLTTDEEIYETLRN VVTHGAHPVGTAAMSHPDASYGVVDPDLKVKGVQGLRVVDASIMPYLIVGHTQAPVYAIAERAADLIKASW |
| Length | 596 |
Coding
| Sequence id | CopciAB_435561.T0 |
|---|---|
| Sequence |
>CopciAB_435561.T0 ATGGGTACCAGGTCGCTTCTTCCTTCTCTCCTCGTCCTTGCTTCTTTCCTCTTCCATCCGTCACTCGCGGTCATT CGAGATGACTTCACCGACTTGCCACGGCTTGCTTACGACTTTGTGATCGTTGGAGGCGGAACCGCTGGCTCCGTC CTAGCAAGCAGGCTGACCGAGGACCCCGACTTCAATGTTTTGCTGATTGAAGCTGGGCCTTCGCACGAGGGTGTG CTCGAATCGCAGGTTCCCGGCTTAACTTTTGCTCTTCAGCAAACGCAATACGATTGGAACTTCAACACCGTTCCT CAGATTGGTCTGAATAACCGGACGGATCGGTTGCCCAGAGGTCGTATGCTAGGTGGAAGTAGCTCCATCAACGGA CTATTCTACACTCGTGGATCTTCGGATGACTATGACAGATGGGCAGCCATCACGGGCGACTCTGGTTGGTCTTGG AGCAACATCCTTCCCTATCTCCTGAAGAGCGAGAAATGGAACCCTCCGACGGACGGTCACGATACCACCGGTAAG TATGATCCCAGCTACCATTCGACCACCGGAAAAGTCGGCGTCAGTCTGTCCGGTGTACCTCAAGCTTCGGATAGC AAGGTCTTCGAAGCTAGCCAGGAGCTTGGTGGGGAGTTTGCGTACGACGTCGACATGAACGACGGAACACCACTT GGTTTTGGTTGGCTTCAGTCCACGATCAACAATGGGGAGAGAAGTAGTGCTGCCACTGGTTACCTCGATGCAACC GTTCTTGCTCGACCGAACCTTCACGTCCTGGTGAACAACCGCGTCGTCAAGTTGGCTGAAAGAAAGCAGCCTGGA TCTAACACCCCAGCGTTCCAAACTGTCCATTTTCAGACAGGAGGCAGTCCTTCAGGCAGGCTACACCGCGTAACT GCCAAGAAGGAAGTAATCCTGGCAGCGGGCGCCTATGGAAGCCCTCAACTGCTCATGTTATCCGGGATTGGTGAT TCGACCGAGCTCCAGAACATCGGTATCGAGCCCAAAGTGAAACTTCCTAGCGTCGGCAAGAACCTCACTGAACAC GTCCAATTCAGCATGATGTTCAGCTTGGGCATAAACGACACCGTCGACCCATTCCTGAACCAAACCTTCCTCATG GAAAGCATGCAGGAGTGGTTCGACCATCGTACCGGATATATGACGAACCTAGGTATCAATTCTGTTATCTGGCAC AGACTCCCCGAAGACTGGCCTCACTGGAGTCAGTTTGAAGACCCTACGGGTGGTATTAATTCGCCGCATATTGAG TTGGTCGTGATGGGTGGTGGGATTTACCCCACTCCTGGACCTAATCTCATTGTCAGCCCTGTCCTGGCGAGTCCT CGCTCTCGCGGCTTCATGACTCTCAACTCGACTAGCCCATACGCCAGTCCTCTCATCGACCCAGCGATGCTCACC CACCCCTTCGACATCCTAGCTCTGCGCGAGTCTGTCCGCTCTGTTTTGAAATTCGTCGGTGCCTCCACGTGGGAC GAGTACAACCTTACGCTCCTTCCTCCGTTTGCGGGACTTACGACTGACGAAGAAATCTACGAGACCCTTCGCAAC GTTGTTACCCATGGTGCGCATCCTGTCGGCACGGCAGCGATGTCTCATCCAGACGCTTCGTATGGGGTTGTCGAT CCTGATCTGAAGGTCAAGGGAGTTCAGGGCCTTCGTGTAGTCGATGCTTCCATTATGCCCTACCTCATTGTTGGA CATACACAGGCACCTGTGTATGCAATCGCCGAGCGAGCTGCAGATCTCATCAAGGCATCGTGGTAG |
| Length | 1791 |
Transcript
| Sequence id | CopciAB_435561.T0 |
|---|---|
| Sequence |
>CopciAB_435561.T0 AGAGGATCTTGGGAGGCCCCTTCATTGGCAGCCTTCTTCATATCTTCTTCTGCACAGCTACGGTACTATTACCCC GTCACGCCCGCATCTGATGGGTACCAGGTCGCTTCTTCCTTCTCTCCTCGTCCTTGCTTCTTTCCTCTTCCATCC GTCACTCGCGGTCATTCGAGATGACTTCACCGACTTGCCACGGCTTGCTTACGACTTTGTGATCGTTGGAGGCGG AACCGCTGGCTCCGTCCTAGCAAGCAGGCTGACCGAGGACCCCGACTTCAATGTTTTGCTGATTGAAGCTGGGCC TTCGCACGAGGGTGTGCTCGAATCGCAGGTTCCCGGCTTAACTTTTGCTCTTCAGCAAACGCAATACGATTGGAA CTTCAACACCGTTCCTCAGATTGGTCTGAATAACCGGACGGATCGGTTGCCCAGAGGTCGTATGCTAGGTGGAAG TAGCTCCATCAACGGACTATTCTACACTCGTGGATCTTCGGATGACTATGACAGATGGGCAGCCATCACGGGCGA CTCTGGTTGGTCTTGGAGCAACATCCTTCCCTATCTCCTGAAGAGCGAGAAATGGAACCCTCCGACGGACGGTCA CGATACCACCGGTAAGTATGATCCCAGCTACCATTCGACCACCGGAAAAGTCGGCGTCAGTCTGTCCGGTGTACC TCAAGCTTCGGATAGCAAGGTCTTCGAAGCTAGCCAGGAGCTTGGTGGGGAGTTTGCGTACGACGTCGACATGAA CGACGGAACACCACTTGGTTTTGGTTGGCTTCAGTCCACGATCAACAATGGGGAGAGAAGTAGTGCTGCCACTGG TTACCTCGATGCAACCGTTCTTGCTCGACCGAACCTTCACGTCCTGGTGAACAACCGCGTCGTCAAGTTGGCTGA AAGAAAGCAGCCTGGATCTAACACCCCAGCGTTCCAAACTGTCCATTTTCAGACAGGAGGCAGTCCTTCAGGCAG GCTACACCGCGTAACTGCCAAGAAGGAAGTAATCCTGGCAGCGGGCGCCTATGGAAGCCCTCAACTGCTCATGTT ATCCGGGATTGGTGATTCGACCGAGCTCCAGAACATCGGTATCGAGCCCAAAGTGAAACTTCCTAGCGTCGGCAA GAACCTCACTGAACACGTCCAATTCAGCATGATGTTCAGCTTGGGCATAAACGACACCGTCGACCCATTCCTGAA CCAAACCTTCCTCATGGAAAGCATGCAGGAGTGGTTCGACCATCGTACCGGATATATGACGAACCTAGGTATCAA TTCTGTTATCTGGCACAGACTCCCCGAAGACTGGCCTCACTGGAGTCAGTTTGAAGACCCTACGGGTGGTATTAA TTCGCCGCATATTGAGTTGGTCGTGATGGGTGGTGGGATTTACCCCACTCCTGGACCTAATCTCATTGTCAGCCC TGTCCTGGCGAGTCCTCGCTCTCGCGGCTTCATGACTCTCAACTCGACTAGCCCATACGCCAGTCCTCTCATCGA CCCAGCGATGCTCACCCACCCCTTCGACATCCTAGCTCTGCGCGAGTCTGTCCGCTCTGTTTTGAAATTCGTCGG TGCCTCCACGTGGGACGAGTACAACCTTACGCTCCTTCCTCCGTTTGCGGGACTTACGACTGACGAAGAAATCTA CGAGACCCTTCGCAACGTTGTTACCCATGGTGCGCATCCTGTCGGCACGGCAGCGATGTCTCATCCAGACGCTTC GTATGGGGTTGTCGATCCTGATCTGAAGGTCAAGGGAGTTCAGGGCCTTCGTGTAGTCGATGCTTCCATTATGCC CTACCTCATTGTTGGACATACACAGGCACCTGTGTATGCAATCGCCGAGCGAGCTGCAGATCTCATCAAGGCATC GTGGTAGACTTGGAACAAGTAAATGTATATTATGTTATATTTACAAGAACAGGTTTTCGTTACATCGAA |
| Length | 1944 |
Gene
| Sequence id | CopciAB_435561.T0 |
|---|---|
| Sequence |
>CopciAB_435561.T0 AGAGGATCTTGGGAGGCCCCTTCATTGGCAGCCTTCTTCATATCTTCTTCTGCACAGCTACGGTACTATTACCCC GTCACGCCCGCATCTGATGGGTACCAGGTCGCTTCTTCCTTCTCTCCTCGTCCTTGCTTCTTTCCTCTTCCATCC GTCACTCGCGGTCATTCGAGATGACTTCACCGACTTGCCACGGCTTGCTTACGACTTTGTGATCGTTGGAGGTAC GTATTTCTACTTTACCTCATTGAAGGAACTCTCTTTCGGAATGCTGATAAACATCTCTAGGCGGAACCGCTGGCT CCGTCCTAGCAAGCAGGCTGACCGAGGACCCCGACTTCAATGTTTTGCTGATTGAAGCTGGGCCTTCGTAAGTCC TCTCAGTTGCGATCTATCTTTCGTGGGCGCTGACGATGGCCATAGGCACGAGGGTGTGCTCGAATCGCAGGTTCC CGGCTTAACTTTTGCTCTTCAGCAAACGCAATACGATTGGAACTTCAACACCGTTCCTCAGATTGGTCTGAATAA CCGGACGGATCGGTTGCCCAGAGGTCGTATGCTAGGTGGAAGTAGCTCCATCAGTACGCGCTGCCATTCCCTCTT GTGCATTCTTGGGCTCTGACCTGTCCTACACAGACGGACTATTCTACACTCGTGGATCTTCGGATGACTATGACA GATGGGCAGCCATCACGGGCGACTCTGGTTGGTCTTGGAGCAACATCCTTCCCTATCTCCTGAAGGTCAGCCCAT CCCTAGAGTCTGAGAGCCAGCCGGGGCTGAGATTGGTGCGAATAGAGCGAGAAATGGAACCCTCCGACGGACGGT CACGATACCACCGGTAAGTATGATCCCAGCTACCATTCGACCACCGGAAAAGTCGGCGTCAGTCTGTCCGGTGTA CCTCAAGCTTCGGATAGCAAGGTCTTCGAAGCTAGCCAGGAGCTTGGTGGGGAGTTTGCGTACGACGTCGACATG AACGACGGAACACCACTTGGTTTTGGTACGAATATTGCTTCCCTTTTTCTCGGGAGTGACATCTGAGAGATTTCG ATTTAGGTTGGCTTCAGTCCACGATCAACAATGGGGAGAGAAGTAGTGCTGCCACTGGTTACCTCGATGCAACCG TTCTTGCTCGACCGAACCTTCACGTCCTGGTGAACAACCGCGTCGTCAAGTTGGCTGAAAGAAAGCAGCCTGGAT CTAACACCCCAGCGTTCCAAACTGTCCATTTTCAGACAGGAGGCAGTCCTTCAGGTAAGCACATCCTCCCTACTC AATGCCCAATTGAGTGTTAAACCGTCTTTCCACAGGCAGGCTACACCGCGTAACTGCCAAGAAGGAAGTAATCCT GGCAGCGGGCGCCTATGGAAGCCCTCAACTGCTCATGTTATCCGGGATTGGTGATTCGACCGAGCTCCAGAACAT CGGTATCGAGCCCAAAGTGAAACTTCCTAGCGTCGGCAAGAACCTCACTGAACACGTCCAATTCAGCATGATGTT CAGCTTGGGCATAAACGACACCGTCGACCCATTCCTGAACCAAACCTTCCTCATGGAAAGCATGCAGGAGTGGTT CGACCATCGTACCGGATATATGACGAACCTAGGTATCAATTCTGTTATCTGGCACAGACTCCCCGAAGACTGGCC TCACTGGAGTCAGTTTGAAGACCCTACGGGTGGTATTAATTCGCCGCATATTGAGTTGGTCGTGATGGGTGGTGG GATTTACCCCACTCCTGGACCTAATCTCATTGTCAGCCCTGTCCTGGCGAGTCCTCGCTCTCGTACGCCCCATTC AAGCATACTCAACGCTGAACCCATTCGCTGACCATTCACTCCTCTAGGCGGCTTCATGACTCTCAACTCGACTAG CCCATACGCCAGTCCTCTCATCGACCCAGCGATGCTCACCCACCCCTTCGACATCCTAGCTCTGCGCGAGTCTGT CCGCTCTGTTTTGAAATTCGTCGGTGCCTCCACGTGGGACGAGTACAACCTTACGCTCCTTCCTCCGTTTGCGGG ACTTACGACTGACGAAGAAATCTACGAGACCCTTCGCAACGTTGTTACCCATGGTGCGCATCCTGTCGGCACGGC AGCGATGTCTCATCCAGACGCTTCGTATGGGGTTGTCGATCCTGATCTGAAGGTCAAGGGAGTTCAGGGCCTTCG TGTAGTCGATGCTTCCATTATGGTGGGCTTCTTCCCTGGTTAATTCCATTTTAAATTAGACTGACGAGGTCTTTC CAGCCCTACCTCATTGTTGGACATACACAGGCACCTGTGTATGCAATCGCCGAGCGAGCTGCAGATCTCATCAAG GCATCGTGGTAGACTTGGAACAAGTAAATGTATATTATGTTATATTTACAAGAACAGGTTTTCGTTACATCGAA |
| Length | 2399 |