CopciAB_439087
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_439087 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 439087 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_2:2946275..2949835 | Strand | - |
| Gene length (nt) | 3561 | Transcript length (nt) | 3505 |
| CDS length (nt) | 2568 | Protein length (aa) | 855 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7265270 | 38.1 | 9.562E-166 | 542 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_118488 | 38.4 | 9.675E-160 | 524 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_183603 | 41.9 | 2.366E-156 | 514 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1110814 | 34.2 | 1.865E-127 | 429 |
| Pleurotus ostreatus PC9 | PleosPC9_1_68860 | 34.1 | 8.898E-126 | 424 |
| Lentinula edodes B17 | Lened_B_1_1_2966 | 31.3 | 4.082E-124 | 419 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_11892 | 31.3 | 5.119E-124 | 419 |
| Lentinula edodes NBRC 111202 | Lenedo1_1166228 | 31.2 | 1.181E-121 | 412 |
| Flammulina velutipes | Flave_chr09AA00337 | 36.2 | 2.69E-115 | 393 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_132780 | 35.5 | 6.238E-110 | 377 |
| Grifola frondosa | Grifr_OBZ75982 | 31.9 | 4.107E-79 | 284 |
| Pleurotus eryngii ATCC 90797 | Pleery1_834833 | 42.7 | 7.967E-65 | 240 |
| Auricularia subglabra | Aurde3_1_1324498 | 27.9 | 2.619E-51 | 198 |
| Schizophyllum commune H4-8 | Schco3_2004633 | 29.6 | 5.434E-32 | 135 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_439087.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 427 | 769 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR000719 | Protein kinase domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
KEGG
| KEGG Orthology |
|---|
| K14864 |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
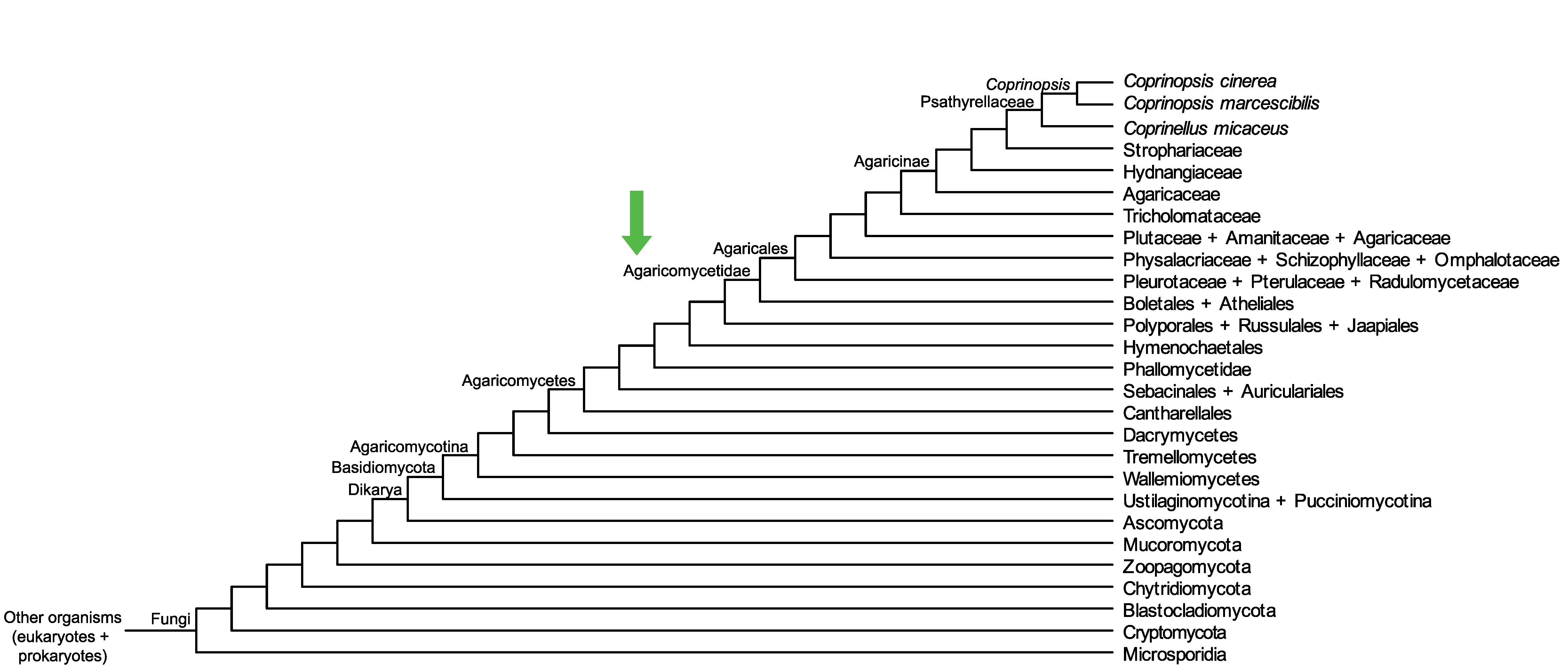
Conservation of CopciAB_439087 across fungi.
Arrow shows the origin of gene family containing CopciAB_439087.
Protein
| Sequence id | CopciAB_439087.T0 |
|---|---|
| Sequence |
>CopciAB_439087.T0 MSTSSASTRSTSSSFSSTFSDVDDPMFSPPSSPPASSKTHAFFASSFAASPPPPRPPLSLKKSDNKPTAVKNRDD NGTNPPPAASPGLFSSAKTSRFNVSRIFPSRYARSRRDSAEQQDVQNAFSSNALRRKPDFLFIDVDGDADPNEIH HIDHRETPFKDVFLSLPTAPSYHDRAGSDSSLSPPSSISSSSSHAEIRLPTPPPPPPLLTASSNASDNEFTPRPA DLEPSIAAVQPQELSPLSPLSEDNSIPADTELHPGDVIADSSFASATSPSLRLLRPLGQGAFSAVWLAEDLSSIP LVLSSKRSVRDLKRQASLQSKGSVSRKSSLKSPTSSRGVSRNSSMKRFMARVTGTQPSVDSLLLDGHVHHEGRET TWTSPDGHLHPMPPSSMHPSPSPSSSISSLPSPEVGLGASLSRQSSTSTRRSATSRAEARVVAVKLTPRRPEPNS PLTEERTRVGFIREVEVLRHISHPNITPLLAHFSTPTYHVLVLPYLKGGDLLSLVNNDAVWGRLSESVLRRIWCE ICRAVGWMHSVGIVHRDIKLENVLLTWDGLQSYSNLISTELSSTPTESSILSIPRPTVADLPTPIVKLTDFGLSR FIDIGNGERGQGEMLNTRCGSEAYAAPELVMGGGPRGVYDGRETDAWACGVVLYALVGRKLPFGEGVGIGMDGGH GTQSHINGERVHPPGYSSRKHAVERRAWLMRIAKGEYEWPEVPSDAAAVEDGDGELVGPRLALSEGAKRVVGRLL VRDPKKRARIVDLWEDDWMRGDEIGLGMWEAIQLDRNASDRVSDGSGEVKPATNENPITVLKEDGLDDGEWEPLD EDEYLEEEDEHEGWLLDEQSISSIVRQEVV |
| Length | 855 |
Coding
| Sequence id | CopciAB_439087.T0 |
|---|---|
| Sequence |
>CopciAB_439087.T0 ATGTCCACATCCTCGGCGTCGACTCGGTCCACGTCCTCCTCATTCTCCTCTACCTTTTCAGATGTCGACGACCCA ATGTTCAGCCCGCCCTCGTCGCCCCCAGCTTCCTCAAAGACCCATGCCTTTTTTGCTTCCTCCTTCGCCGCCAGC CCGCCGCCGCCCCGCCCACCTCTCTCTTTGAAGAAGTCCGACAACAAGCCCACCGCCGTCAAGAACAGGGACGAC AATGGCACAAACCCTCCACCAGCAGCGAGCCCGGGTCTCTTCTCGTCCGCCAAAACATCGCGCTTCAATGTTTCA CGGATCTTTCCCTCAAGATATGCCCGCTCTCGCCGTGACTCTGCAGAGCAGCAAGACGTCCAGAACGCATTCTCC TCGAACGCCCTGCGCAGAAAACCCGACTTTCTCTTCATAGACGTCGACGGAGATGCAGACCCGAATGAAATACAT CACATTGACCATAGAGAGACTCCATTCAAGGACGTGTTCCTCTCTCTTCCCACAGCCCCGTCCTACCACGACCGC GCAGGATCAGACTCGTCGCTCTCCCCACCCTCGTCGATATCTTCGTCGTCTTCGCACGCAGAGATTCGGCTACCC ACCCCGCCGCCGCCCCCGCCCCTCCTAACCGCAAGTTCCAACGCCTCAGACAATGAATTCACACCCCGGCCAGCT GACCTCGAACCTTCCATAGCCGCTGTACAGCCCCAGGAACTTTCTCCCTTATCGCCTCTCAGCGAAGACAACAGT ATCCCAGCCGACACCGAATTGCACCCTGGTGATGTCATAGCCGACTCCAGTTTCGCCTCGGCTACTTCCCCGTCC CTGCGCCTCCTCCGTCCTCTCGGCCAGGGCGCCTTTTCCGCAGTATGGCTTGCAGAAGACCTGTCGTCTATACCC CTCGTTCTCTCCTCGAAGAGGAGCGTCCGTGACCTCAAGCGACAGGCGAGCCTGCAATCGAAAGGAAGTGTCTCG CGCAAGTCGAGTCTGAAAAGCCCAACGTCGAGCAGGGGTGTTTCCCGCAACTCGAGTATGAAGAGGTTCATGGCA CGCGTCACTGGCACACAGCCTTCGGTCGATAGCCTGTTGTTGGACGGCCATGTCCACCATGAAGGGCGGGAAACT ACTTGGACCAGTCCAGATGGTCACCTCCACCCCATGCCCCCGTCGTCAATGCACCCTAGTCCATCACCATCATCC TCAATATCATCTCTTCCTTCTCCCGAGGTCGGGTTGGGTGCATCGTTGTCACGCCAAAGCTCCACAAGTACACGC CGCTCGGCTACTTCTCGGGCAGAGGCCCGAGTAGTCGCGGTCAAGCTGACACCCCGGCGTCCGGAGCCAAACTCT CCTTTGACTGAAGAGAGGACCAGGGTGGGATTCATTCGTGAAGTCGAGGTACTCCGACACATCTCTCATCCCAAC ATCACGCCCTTGCTGGCCCACTTCTCCACTCCGACATATCACGTCCTCGTGCTGCCCTACCTCAAGGGCGGGGAT CTGTTGTCGCTTGTCAATAATGATGCCGTTTGGGGTAGACTGTCTGAGAGTGTGTTAAGGCGGATATGGTGTGAG ATATGCCGTGCGGTCGGGTGGATGCATAGCGTCGGAATCGTGCACCGCGACATCAAGTTGGAGAATGTCCTGTTG ACCTGGGATGGCCTCCAGTCGTATTCCAACCTCATCTCGACCGAGCTCTCGTCGACGCCTACCGAATCGTCGATA TTGTCAATACCACGACCCACCGTGGCTGATCTCCCCACCCCCATTGTCAAACTCACCGACTTTGGTCTGTCGCGG TTCATTGACATCGGCAATGGCGAACGCGGGCAGGGCGAGATGCTCAATACCCGCTGCGGATCAGAAGCCTATGCC GCACCTGAATTGGTTATGGGTGGCGGTCCACGAGGCGTTTATGACGGACGCGAAACGGATGCGTGGGCCTGTGGT GTAGTCTTGTATGCGCTTGTGGGTCGCAAGCTTCCTTTCGGTGAAGGTGTTGGAATTGGTATGGACGGTGGACAC GGCACACAGAGCCATATCAATGGCGAACGTGTCCACCCTCCTGGCTACTCTAGCCGGAAGCATGCCGTGGAGCGT CGTGCGTGGTTGATGAGGATTGCGAAGGGCGAGTACGAGTGGCCTGAAGTCCCGTCTGATGCAGCTGCGGTAGAG GATGGGGATGGGGAACTCGTTGGGCCTCGGTTGGCGTTGAGCGAGGGTGCGAAGAGGGTTGTTGGGCGGTTGTTG GTTCGTGATCCGAAGAAGAGGGCTAGGATTGTGGATCTGTGGGAGGATGACTGGATGCGCGGGGATGAGATTGGG TTGGGGATGTGGGAAGCTATCCAGCTCGATCGTAATGCCTCGGATCGTGTCTCGGATGGCTCTGGGGAGGTGAAG CCTGCGACAAACGAGAACCCCATCACCGTGCTCAAGGAGGACGGGCTCGATGATGGGGAGTGGGAGCCTCTTGAC GAAGATGAATATCTCGAAGAGGAGGATGAACATGAAGGATGGCTGTTGGATGAGCAGAGTATCTCCAGTATAGTG AGGCAAGAAGTTGTGTAG |
| Length | 2568 |
Transcript
| Sequence id | CopciAB_439087.T0 |
|---|---|
| Sequence |
>CopciAB_439087.T0 AGCTTTCTCTCTGATCTCACATGTCCACATCCTCGGCGTCGACTCGGTCCACGTCCTCCTCATTCTCCTCTACCT TTTCAGATGTCGACGACCCAATGTTCAGCCCGCCCTCGTCGCCCCCAGCTTCCTCAAAGACCCATGCCTTTTTTG CTTCCTCCTTCGCCGCCAGCCCGCCGCCGCCCCGCCCACCTCTCTCTTTGAAGAAGTCCGACAACAAGCCCACCG CCGTCAAGAACAGGGACGACAATGGCACAAACCCTCCACCAGCAGCGAGCCCGGGTCTCTTCTCGTCCGCCAAAA CATCGCGCTTCAATGTTTCACGGATCTTTCCCTCAAGATATGCCCGCTCTCGCCGTGACTCTGCAGAGCAGCAAG ACGTCCAGAACGCATTCTCCTCGAACGCCCTGCGCAGAAAACCCGACTTTCTCTTCATAGACGTCGACGGAGATG CAGACCCGAATGAAATACATCACATTGACCATAGAGAGACTCCATTCAAGGACGTGTTCCTCTCTCTTCCCACAG CCCCGTCCTACCACGACCGCGCAGGATCAGACTCGTCGCTCTCCCCACCCTCGTCGATATCTTCGTCGTCTTCGC ACGCAGAGATTCGGCTACCCACCCCGCCGCCGCCCCCGCCCCTCCTAACCGCAAGTTCCAACGCCTCAGACAATG AATTCACACCCCGGCCAGCTGACCTCGAACCTTCCATAGCCGCTGTACAGCCCCAGGAACTTTCTCCCTTATCGC CTCTCAGCGAAGACAACAGTATCCCAGCCGACACCGAATTGCACCCTGGTGATGTCATAGCCGACTCCAGTTTCG CCTCGGCTACTTCCCCGTCCCTGCGCCTCCTCCGTCCTCTCGGCCAGGGCGCCTTTTCCGCAGTATGGCTTGCAG AAGACCTGTCGTCTATACCCCTCGTTCTCTCCTCGAAGAGGAGCGTCCGTGACCTCAAGCGACAGGCGAGCCTGC AATCGAAAGGAAGTGTCTCGCGCAAGTCGAGTCTGAAAAGCCCAACGTCGAGCAGGGGTGTTTCCCGCAACTCGA GTATGAAGAGGTTCATGGCACGCGTCACTGGCACACAGCCTTCGGTCGATAGCCTGTTGTTGGACGGCCATGTCC ACCATGAAGGGCGGGAAACTACTTGGACCAGTCCAGATGGTCACCTCCACCCCATGCCCCCGTCGTCAATGCACC CTAGTCCATCACCATCATCCTCAATATCATCTCTTCCTTCTCCCGAGGTCGGGTTGGGTGCATCGTTGTCACGCC AAAGCTCCACAAGTACACGCCGCTCGGCTACTTCTCGGGCAGAGGCCCGAGTAGTCGCGGTCAAGCTGACACCCC GGCGTCCGGAGCCAAACTCTCCTTTGACTGAAGAGAGGACCAGGGTGGGATTCATTCGTGAAGTCGAGGTACTCC GACACATCTCTCATCCCAACATCACGCCCTTGCTGGCCCACTTCTCCACTCCGACATATCACGTCCTCGTGCTGC CCTACCTCAAGGGCGGGGATCTGTTGTCGCTTGTCAATAATGATGCCGTTTGGGGTAGACTGTCTGAGAGTGTGT TAAGGCGGATATGGTGTGAGATATGCCGTGCGGTCGGGTGGATGCATAGCGTCGGAATCGTGCACCGCGACATCA AGTTGGAGAATGTCCTGTTGACCTGGGATGGCCTCCAGTCGTATTCCAACCTCATCTCGACCGAGCTCTCGTCGA CGCCTACCGAATCGTCGATATTGTCAATACCACGACCCACCGTGGCTGATCTCCCCACCCCCATTGTCAAACTCA CCGACTTTGGTCTGTCGCGGTTCATTGACATCGGCAATGGCGAACGCGGGCAGGGCGAGATGCTCAATACCCGCT GCGGATCAGAAGCCTATGCCGCACCTGAATTGGTTATGGGTGGCGGTCCACGAGGCGTTTATGACGGACGCGAAA CGGATGCGTGGGCCTGTGGTGTAGTCTTGTATGCGCTTGTGGGTCGCAAGCTTCCTTTCGGTGAAGGTGTTGGAA TTGGTATGGACGGTGGACACGGCACACAGAGCCATATCAATGGCGAACGTGTCCACCCTCCTGGCTACTCTAGCC GGAAGCATGCCGTGGAGCGTCGTGCGTGGTTGATGAGGATTGCGAAGGGCGAGTACGAGTGGCCTGAAGTCCCGT CTGATGCAGCTGCGGTAGAGGATGGGGATGGGGAACTCGTTGGGCCTCGGTTGGCGTTGAGCGAGGGTGCGAAGA GGGTTGTTGGGCGGTTGTTGGTTCGTGATCCGAAGAAGAGGGCTAGGATTGTGGATCTGTGGGAGGATGACTGGA TGCGCGGGGATGAGATTGGGTTGGGGATGTGGGAAGCTATCCAGCTCGATCGTAATGCCTCGGATCGTGTCTCGG ATGGCTCTGGGGAGGTGAAGCCTGCGACAAACGAGAACCCCATCACCGTGCTCAAGGAGGACGGGCTCGATGATG GGGAGTGGGAGCCTCTTGACGAAGATGAATATCTCGAAGAGGAGGATGAACATGAAGGATGGCTGTTGGATGAGC AGAGTATCTCCAGTATAGTGAGGCAAGAAGTTGTGTAGTAACGCTGGTCTGCACTTTTTCACGACATCCCATCCC ACCGCGCAGCCCCATCCCCGCATCGTTTCGGACCCCGAGATGTATTCCATCATTCGCCACCCGTGTATCTTTATC TTTTAATACTTGTTTCTCCTTTTTCTTGCGGTACCTTACACCACCACAACACCACCACAACCACCACCACCACCA CCACCACCGCCCCCACCGTTCCTTTATTTATTTCCTTTGTTCTCTCCTCTCTCGCTCGCTCGCAACTTACAACCC TTCATTCATCCGTCTCTGTACTCCGATCCTCCTGTGTATTCATATGCTCAACTCCTGTCATCCCGCTTTCTTGTT GTGTTCTTCTGTTCCTCCTTTTTTTTTCCTCCTCCAAACGTGCATCGCTTATTAATTATTCCACCATCGCACGCA ATCTTCCCTCTCTTTCTCGCACAATTTCCCTTTTCCAGAAACAATCTTCCATTTTCAAACCTTTTTTCTTTGTCG TTGTCGCAATTACCGTCTCTCCCAGTCCGTTTGTTGTACATCGTCGCTCGCTGTGTAAAAACAGATGTCGAAAAC CCCCTTTTTTGTTTGTCTGTATACAAAGCATACATATGCGTAGTCGTTTTTTTTCTGTGTTCACTCGTGTAGACG TACGTACGTAGTAGTGTGTACTACCGACCTCTAGTATTTAAAAAGACAATCACACACCCCACCTGCCCGTTTCCA ATTTGTAATGAAAAATGTATGTTTTTTTGTTTATAATAATTTTTATTTGTGTTTTGTGTAGAATATGCCTTGTGT TCTGCTTTGTAGTCGTTTTTTTTTGGTTTCGGCTGTCTTTGTAGAATGTCGTTGGTTTTAATCTTTCTTTTTATT AGAGTCTGTTTTTGTGTACTACATACTTTATCGTTTATATATTGGGTTGGTTGTG |
| Length | 3505 |
Gene
| Sequence id | CopciAB_439087.T0 |
|---|---|
| Sequence |
>CopciAB_439087.T0 AGCTTTCTCTCTGATCTCACATGTCCACATCCTCGGCGTCGACTCGGTCCACGTCCTCCTCATTCTCCTCTACCT TTTCAGATGTCGACGACCCAATGTTCAGCCCGCCCTCGTCGCCCCCAGCTTCCTCAAAGACCCATGCCTTTTTTG CTTCCTCCTTCGCCGCCAGCCCGCCGCCGCCCCGCCCACCTCTCTCTTTGAAGAAGTCCGACAACAAGCCCACCG CCGTCAAGAACAGGGACGACAATGGCACAAACCCTCCACCAGCAGCGAGCCCGGGTCTCTTCTCGTCCGCCAAAA CATCGCGCTTCAATGTTTCACGGATCTTTCCCTCAAGATATGCCCGCTCTCGCCGTGACTCTGCAGAGCAGCAAG ACGTCCAGAACGCATTCTCCTCGAACGCCCTGCGCAGAAAACCCGACTTTCTCTTCATAGACGTCGACGGAGATG CAGACCCGAATGAAATACATCACATTGACCATAGAGAGACTCCATTCAAGGACGTGTTCCTCTCTCTTCCCACAG CCCCGTCCTACCACGACCGCGCAGGATCAGACTCGTCGCTCTCCCCACCCTCGTCGATATCTTCGTCGTCTTCGC ACGCAGAGATTCGGCTACCCACCCCGCCGCCGCCCCCGCCCCTCCTAACCGCAAGTTCCAACGCCTCAGACAATG AATTCACACCCCGGCCAGCTGACCTCGAACCTTCCATAGCCGCTGTACAGCCCCAGGAACTTTCTCCCTTATCGC CTCTCAGCGAAGACAACAGTATCCCAGCCGACACCGAATTGCACCCTGGTGATGTCATAGCCGACTCCAGTTTCG CCTCGGCTACTTCCCCGTCCCTGCGCCTCCTCCGTCCTCTCGGCCAGGGCGCCTTTTCCGCAGTATGGCTTGCAG AAGACCTGTCGTCTATACCCCTCGTTCTCTCCTCGAAGAGGAGCGTCCGTGACCTCAAGCGACAGGCGAGCCTGC AATCGAAAGGAAGTGTCTCGCGCAAGTCGAGTCTGAAAAGCCCAACGTCGAGCAGGGGTGTTTCCCGCAACTCGA GTATGAAGAGGTTCATGGCACGCGTCACTGGCACACAGCCTTCGGTCGATAGCCTGTTGTTGGACGGCCATGTCC ACCATGAAGGGCGGGAAACTACTTGGACCAGTCCAGATGGTCACCTCCACCCCATGCCCCCGTCGTCAATGCACC CTAGTCCATCACCATCATCCTCAATATCATCTCTTCCTTCTCCCGAGGTCGGGTTGGGTGCATCGTTGTCACGCC AAAGCTCCACAAGTACACGCCGCTCGGCTACTTCTCGGGCAGAGGCCCGAGTAGTCGCGGTCAAGCTGACACCCC GGCGTCCGGAGCCAAACTCTCCTTTGACTGAAGAGAGGACCAGGGTGGGATTCATTCGTGAAGTCGAGGTACTCC GACACATCTCTCATCCCAACATCACGCCCTTGCTGGCCCACTTCTCCACTCCGACATATCACGTCCTCGTGCTGC CCTACCTCAAGGGCGGGGATCTGTTGTCGCTTGTCAATAATGATGCCGTTTGGGGTAGACTGTCTGAGAGTGTGT TAAGGCGGATATGGTGTGAGATATGCCGTGCGGTCGGGTGGATGCATAGCGTCGGAATCGTGCACCGCGACATCA AGTTGGAGAATGTCCTGTTGACCTGGGATGGCCTCCAGTCGTATTCCAACCTCATCTCGACCGAGCTCTCGTCGA CGCCTACCGAATCGTCGATATTGTCAATACCACGACCCACCGTGGCTGATCTCCCCACCCCCATTGTCAAACTCA CCGACTTTGGTCTGTCGCGGTTCATTGACATCGGCAATGGCGAACGCGGGCAGGGCGAGATGCTCAATACCCGCT GCGGATCAGAAGCCTATGCCGCACCTGAATTGGTTATGGGTGGCGGTCCACGAGGCGTTTATGACGGACGCGAAA CGGATGCGTGGGCCTGTGGTGTAGTCTTGTATGCGCTTGTGGGTCGCAAGCTTCCTTTCGGTGAAGGTGTTGGAA TTGGTATGGACGGTGGACACGGCACACAGAGCCATATCAATGGCGAACGTGTCCACCCTCCTGGCTACTCTAGCC GGAAGCATGCCGTGGAGCGTCGTGCGTGGTTGATGAGGATTGCGAAGGGCGAGTACGAGTGGCCTGAAGTCCCGT CTGATGCAGCTGCGGTAGAGGATGGGGATGGGGAACTCGTTGGGCCTCGGTTGGCGTTGAGCGAGGGTGCGAAGA GGGTTGTTGGGCGGTTGTTGGTTCGTGATCCGAAGAAGAGGGCTAGGATTGTGGATCTGTGGGAGGATGACTGGA TGCGCGGGGATGAGATTGGGTTGGGGATGTGGGAAGCTATCCAGCTCGATCGTAATGCCTCGGATCGTGTCTCGG ATGGCTCTGGGGAGGTGAAGCCTGCGACAAACGAGAACCCCATCACCGTGCTCAAGGAGGACGGGCTCGATGATG GGGAGTGGGAGCCTCTTGACGAAGATGAATATCTCGAAGAGGAGGATGAACATGAAGGATGGCTGTTGGATGAGC AGAGTATCTCCAGTATAGTGAGGCAAGAAGTTGTGTAGTAACGCTGGTCTGCACTTTTTGTGAGTGTTTTCATTT TTTCTATCGCCATTGATTTTGATTTCCCCTTCCTGGACAGCACGACATCCCATCCCACCGCGCAGCCCCATCCCC GCATCGTTTCGGACCCCGAGATGTATTCCATCATTCGCCACCCGTGTATCTTTATCTTTTAATACTTGTTTCTCC TTTTTCTTGCGGTACCTTACACCACCACAACACCACCACAACCACCACCACCACCACCACCACCGCCCCCACCGT TCCTTTATTTATTTCCTTTGTTCTCTCCTCTCTCGCTCGCTCGCAACTTACAACCCTTCATTCATCCGTCTCTGT ACTCCGATCCTCCTGTGTATTCATATGCTCAACTCCTGTCATCCCGCTTTCTTGTTGTGTTCTTCTGTTCCTCCT TTTTTTTTCCTCCTCCAAACGTGCATCGCTTATTAATTATTCCACCATCGCACGCAATCTTCCCTCTCTTTCTCG CACAATTTCCCTTTTCCAGAAACAATCTTCCATTTTCAAACCTTTTTTCTTTGTCGTTGTCGCAATTACCGTCTC TCCCAGTCCGTTTGTTGTACATCGTCGCTCGCTGTGTAAAAACAGATGTCGAAAACCCCCTTTTTTGTTTGTCTG TATACAAAGCATACATATGCGTAGTCGTTTTTTTTCTGTGTTCACTCGTGTAGACGTACGTACGTAGTAGTGTGT ACTACCGACCTCTAGTATTTAAAAAGACAATCACACACCCCACCTGCCCGTTTCCAATTTGTAATGAAAAATGTA TGTTTTTTTGTTTATAATAATTTTTATTTGTGTTTTGTGTAGAATATGCCTTGTGTTCTGCTTTGTAGTCGTTTT TTTTTGGTTTCGGCTGTCTTTGTAGAATGTCGTTGGTTTTAATCTTTCTTTTTATTAGAGTCTGTTTTTGTGTAC TACATACTTTATCGTTTATATATTGGGTTGGTTGTG |
| Length | 3561 |