CopciAB_441896
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_441896 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 441896 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_1:3623287..3626783 | Strand | - |
| Gene length (nt) | 3497 | Transcript length (nt) | 2887 |
| CDS length (nt) | 2577 | Protein length (aa) | 858 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7267722 | 32.6 | 3.493E-93 | 327 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB24586 | 29.1 | 6.052E-81 | 290 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_130693 | 27.2 | 1.574E-66 | 245 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_114695 | 27.7 | 1.05E-62 | 233 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1452165 | 25.9 | 1.05E-59 | 224 |
| Lentinula edodes NBRC 111202 | Lenedo1_49119 | 28.1 | 1.936E-58 | 220 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1111154 | 24.9 | 7.134E-58 | 218 |
| Pleurotus ostreatus PC9 | PleosPC9_1_90612 | 25.4 | 2.66E-56 | 213 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_4465 | 26.6 | 2.18E-48 | 188 |
| Grifola frondosa | Grifr_OBZ79374 | 35.1 | 2.268E-36 | 149 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_441896.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 203 | 425 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K13302 |
| K13303 |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
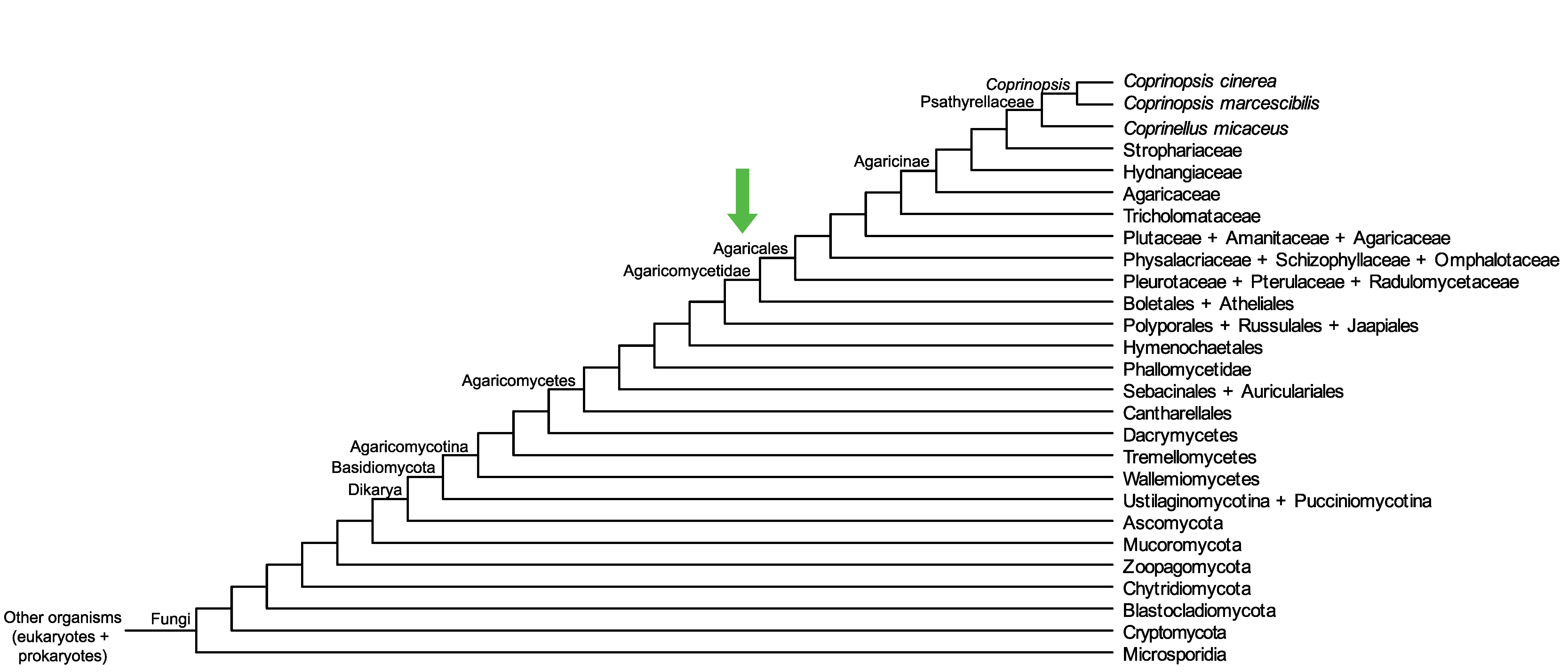
Conservation of CopciAB_441896 across fungi.
Arrow shows the origin of gene family containing CopciAB_441896.
Protein
| Sequence id | CopciAB_441896.T0 |
|---|---|
| Sequence |
>CopciAB_441896.T0 MASDMYRSSFCDFSDSSECTDTSTGTVLRDLSLSPRPSVVDGLGEVSTNSTTPPRLDVEWPNSPPLAVALRFSGI FDDADVAEIEGDLTRQTHQSRSPTFDMTYDIHSFPNIKDINLEMGNDSDISEVVVMSQKQLPTALSTIPEANEPS PVLAYTTSLSDFDIISSYNAPDSDTRVMICQRKRNGNLYKIRSRRIYDHFPWMEQDILETLRDMGAPFLPYLKSV FREENQVHLVLDYWPSASLRELLAKYGTLGSQRVLFYASELLTGIANLHGVGIMHRNLNPDDIILDKRGHIVITN FEFANTIPPSGVNANSFNSDLVLSIPRFHGYQAPEMVLGWAHDISVDCWGFGMLLYFMYFGKHPFQEDRNPVDHR VLRARIIQGSLSPESLRLIHPHTRDIILKCVERNPRIRLNVDGIKNHEYFNNVPWGKVADKLVEVPPLPGFFNDN GSSGELLRRSHSSGLRHTLYQEGAEPVVATPEQPRPALLSPIPSHRPSHPLPLSPSPSAVTFASRPMSPSVLTEM SEPLEGNLSIQSPKPRTFRPFLDPQASAQSAKGGQVDSGMEIWDLLDREEQGSIASSNLGRSHVLGFLRPRKLRK NNSSTSLFRPFVLNLSTSSLTRGLKKKKSRSSTRLGESSATVHDPQQRNQDVELPAGIEHIGSGIGFHRNTLSSD SGHSRSSASSNLPRFCRGGVKLGHRFSFSALRPSGARKIFGRNTRSRSLSSTMATNSGDASALGGLRLAHNGNSP AYVHGHTGALPGSHTSPRTPHEVQASTPILQMNPMSPSGSSSTSASASSSPRTEDGPLTPTTVACDEECQARVSS KAVMDFDEVNKERGRNDLTLRLVPSSRGLQLSV |
| Length | 858 |
Coding
| Sequence id | CopciAB_441896.T0 |
|---|---|
| Sequence |
>CopciAB_441896.T0 ATGGCTTCCGACATGTATCGGTCCTCGTTCTGCGACTTTAGCGATTCATCTGAGTGCACCGATACCTCGACGGGA ACCGTTCTTCGTGATCTCTCCCTCTCACCTCGACCTTCGGTTGTCGACGGACTTGGGGAAGTCTCTACCAACAGT ACAACCCCTCCCCGCCTTGACGTGGAATGGCCCAATTCGCCTCCGTTGGCGGTTGCCCTCAGGTTCTCGGGCATA TTCGACGATGCTGATGTCGCAGAGATCGAAGGAGATCTTACTCGCCAGACTCACCAAAGCAGATCGCCCACTTTC GACATGACCTATGACATTCATAGCTTCCCGAATATCAAGGACATCAACTTGGAGATGGGGAACGACAGTGACATC TCCGAGGTTGTGGTCATGTCGCAGAAACAGCTCCCCACCGCGCTTTCTACGATCCCAGAGGCAAATGAGCCAAGC CCTGTTCTTGCGTATACGACGAGCCTGTCCGACTTTGATATCATCTCCTCTTACAATGCCCCTGATTCTGATACT CGTGTTATGATCTGCCAAAGGAAAAGGAATGGAAACCTTTACAAGATTCGTTCTCGCCGCATCTATGACCATTTT CCCTGGATGGAGCAAGACATCCTGGAGACGCTACGAGATATGGGAGCTCCCTTCCTGCCCTATCTGAAGTCTGTC TTCCGAGAGGAAAATCAAGTGCATCTTGTTCTGGATTATTGGCCTTCAGCGAGTTTGAGAGAGCTCCTCGCAAAG TACGGAACTCTTGGCTCTCAGCGTGTCCTGTTCTATGCTTCTGAACTCCTGACTGGGATTGCCAACCTCCACGGT GTCGGGATCATGCATCGCAACCTTAACCCTGACGATATCATCTTGGACAAGCGGGGGCACATTGTCATTACCAAC TTTGAATTCGCCAACACGATTCCCCCTTCCGGTGTTAACGCCAATTCGTTTAACAGTGATTTGGTTTTGTCGATA CCGAGGTTCCATGGATATCAGGCCCCAGAGATGGTTCTGGGTTGGGCGCATGATATCTCCGTTGATTGCTGGGGA TTTGGGATGTTGCTTTACTTCATGTACTTTGGGAAGCACCCCTTCCAAGAGGACAGAAACCCGGTAGATCATCGC GTCTTGCGCGCACGCATCATTCAAGGCTCTTTGTCTCCAGAGTCTCTCAGGTTGATTCATCCGCACACGAGGGAT ATCATCTTGAAGTGTGTCGAAAGGAATCCGCGTATTCGATTGAATGTCGATGGGATTAAGAACCACGAATATTTT AACAACGTGCCTTGGGGAAAGGTGGCCGATAAGCTCGTCGAAGTCCCACCACTACCCGGGTTCTTCAACGATAAT GGATCCAGTGGGGAGCTTTTGAGAAGGTCCCATTCGTCAGGCTTGCGACACACCCTCTACCAAGAGGGAGCCGAG CCTGTTGTCGCAACACCTGAGCAACCTCGACCTGCGCTACTGAGTCCTATCCCTTCCCATCGTCCTTCTCACCCG CTTCCTCTCTCCCCTTCTCCTTCCGCTGTCACATTCGCCTCTCGTCCCATGTCTCCATCTGTCCTGACTGAGATG TCAGAACCTCTGGAGGGCAATCTCAGCATCCAATCTCCTAAGCCCCGGACGTTTCGACCCTTTCTCGACCCTCAG GCGAGCGCTCAGTCGGCGAAAGGAGGACAAGTAGACTCGGGCATGGAAATCTGGGATCTACTGGATCGTGAGGAG CAAGGCTCGATCGCCTCTTCCAACCTGGGCCGTAGCCACGTTCTTGGATTCCTCAGGCCTCGTAAGCTTCGCAAG AACAATTCGTCGACCAGTCTCTTCAGGCCGTTCGTACTCAACCTGAGCACGTCGTCCTTGACTCGTGGGTTGAAG AAGAAGAAGTCTAGAAGCTCCACTCGCTTGGGCGAGAGTTCCGCTACCGTACATGACCCTCAGCAGCGCAACCAG GACGTTGAGCTCCCAGCCGGAATCGAACATATCGGTAGTGGAATTGGATTCCACCGTAATACTCTCTCCTCTGAT TCAGGCCATTCGAGGTCTTCAGCTTCCTCAAACTTGCCCAGGTTCTGCCGCGGAGGTGTCAAGCTTGGTCATCGA TTCTCATTCAGTGCTTTGCGGCCCAGCGGTGCTCGCAAGATCTTTGGAAGGAATACCCGCTCGAGGTCTCTCTCT TCTACTATGGCGACCAATTCCGGGGATGCTTCTGCACTCGGGGGCCTCCGGTTGGCTCACAATGGGAATAGCCCT GCCTATGTTCACGGTCACACAGGAGCTCTTCCCGGTTCACATACTTCTCCTAGGACGCCTCACGAGGTGCAAGCT AGTACTCCTATCCTCCAAATGAATCCCATGTCACCTAGCGGTTCGTCATCCACCTCCGCTTCTGCGTCATCATCT CCTAGGACCGAGGATGGCCCATTGACTCCAACCACTGTGGCTTGCGACGAGGAGTGTCAAGCCCGAGTATCCTCC AAGGCAGTGATGGACTTTGACGAGGTCAACAAAGAAAGGGGAAGGAATGATTTGACTTTGCGATTGGTTCCCAGT TCCAGAGGTCTCCAGCTATCGGTTTGA |
| Length | 2577 |
Transcript
| Sequence id | CopciAB_441896.T0 |
|---|---|
| Sequence |
>CopciAB_441896.T0 GTTCTCTTCTTCTTGCATTTACCACTTCGACGCGAACGTCCACTTCTGCCTTGGATCTACGTTGCGCGAAACCTT GCTCTCGACCAAGGTCGCAGAGTACCCGTCTCGCCATTTCACTGGCTCTTCAGCGATACCTCTTCTCATCCACAA CCCCGCCTAGCGACGATGGCTTCCGACATGTATCGGTCCTCGTTCTGCGACTTTAGCGATTCATCTGAGTGCACC GATACCTCGACGGGAACCGTTCTTCGTGATCTCTCCCTCTCACCTCGACCTTCGGTTGTCGACGGACTTGGGGAA GTCTCTACCAACAGTACAACCCCTCCCCGCCTTGACGTGGAATGGCCCAATTCGCCTCCGTTGGCGGTTGCCCTC AGGTTCTCGGGCATATTCGACGATGCTGATGTCGCAGAGATCGAAGGAGATCTTACTCGCCAGACTCACCAAAGC AGATCGCCCACTTTCGACATGACCTATGACATTCATAGCTTCCCGAATATCAAGGACATCAACTTGGAGATGGGG AACGACAGTGACATCTCCGAGGTTGTGGTCATGTCGCAGAAACAGCTCCCCACCGCGCTTTCTACGATCCCAGAG GCAAATGAGCCAAGCCCTGTTCTTGCGTATACGACGAGCCTGTCCGACTTTGATATCATCTCCTCTTACAATGCC CCTGATTCTGATACTCGTGTTATGATCTGCCAAAGGAAAAGGAATGGAAACCTTTACAAGATTCGTTCTCGCCGC ATCTATGACCATTTTCCCTGGATGGAGCAAGACATCCTGGAGACGCTACGAGATATGGGAGCTCCCTTCCTGCCC TATCTGAAGTCTGTCTTCCGAGAGGAAAATCAAGTGCATCTTGTTCTGGATTATTGGCCTTCAGCGAGTTTGAGA GAGCTCCTCGCAAAGTACGGAACTCTTGGCTCTCAGCGTGTCCTGTTCTATGCTTCTGAACTCCTGACTGGGATT GCCAACCTCCACGGTGTCGGGATCATGCATCGCAACCTTAACCCTGACGATATCATCTTGGACAAGCGGGGGCAC ATTGTCATTACCAACTTTGAATTCGCCAACACGATTCCCCCTTCCGGTGTTAACGCCAATTCGTTTAACAGTGAT TTGGTTTTGTCGATACCGAGGTTCCATGGATATCAGGCCCCAGAGATGGTTCTGGGTTGGGCGCATGATATCTCC GTTGATTGCTGGGGATTTGGGATGTTGCTTTACTTCATGTACTTTGGGAAGCACCCCTTCCAAGAGGACAGAAAC CCGGTAGATCATCGCGTCTTGCGCGCACGCATCATTCAAGGCTCTTTGTCTCCAGAGTCTCTCAGGTTGATTCAT CCGCACACGAGGGATATCATCTTGAAGTGTGTCGAAAGGAATCCGCGTATTCGATTGAATGTCGATGGGATTAAG AACCACGAATATTTTAACAACGTGCCTTGGGGAAAGGTGGCCGATAAGCTCGTCGAAGTCCCACCACTACCCGGG TTCTTCAACGATAATGGATCCAGTGGGGAGCTTTTGAGAAGGTCCCATTCGTCAGGCTTGCGACACACCCTCTAC CAAGAGGGAGCCGAGCCTGTTGTCGCAACACCTGAGCAACCTCGACCTGCGCTACTGAGTCCTATCCCTTCCCAT CGTCCTTCTCACCCGCTTCCTCTCTCCCCTTCTCCTTCCGCTGTCACATTCGCCTCTCGTCCCATGTCTCCATCT GTCCTGACTGAGATGTCAGAACCTCTGGAGGGCAATCTCAGCATCCAATCTCCTAAGCCCCGGACGTTTCGACCC TTTCTCGACCCTCAGGCGAGCGCTCAGTCGGCGAAAGGAGGACAAGTAGACTCGGGCATGGAAATCTGGGATCTA CTGGATCGTGAGGAGCAAGGCTCGATCGCCTCTTCCAACCTGGGCCGTAGCCACGTTCTTGGATTCCTCAGGCCT CGTAAGCTTCGCAAGAACAATTCGTCGACCAGTCTCTTCAGGCCGTTCGTACTCAACCTGAGCACGTCGTCCTTG ACTCGTGGGTTGAAGAAGAAGAAGTCTAGAAGCTCCACTCGCTTGGGCGAGAGTTCCGCTACCGTACATGACCCT CAGCAGCGCAACCAGGACGTTGAGCTCCCAGCCGGAATCGAACATATCGGTAGTGGAATTGGATTCCACCGTAAT ACTCTCTCCTCTGATTCAGGCCATTCGAGGTCTTCAGCTTCCTCAAACTTGCCCAGGTTCTGCCGCGGAGGTGTC AAGCTTGGTCATCGATTCTCATTCAGTGCTTTGCGGCCCAGCGGTGCTCGCAAGATCTTTGGAAGGAATACCCGC TCGAGGTCTCTCTCTTCTACTATGGCGACCAATTCCGGGGATGCTTCTGCACTCGGGGGCCTCCGGTTGGCTCAC AATGGGAATAGCCCTGCCTATGTTCACGGTCACACAGGAGCTCTTCCCGGTTCACATACTTCTCCTAGGACGCCT CACGAGGTGCAAGCTAGTACTCCTATCCTCCAAATGAATCCCATGTCACCTAGCGGTTCGTCATCCACCTCCGCT TCTGCGTCATCATCTCCTAGGACCGAGGATGGCCCATTGACTCCAACCACTGTGGCTTGCGACGAGGAGTGTCAA GCCCGAGTATCCTCCAAGGCAGTGATGGACTTTGACGAGGTCAACAAAGAAAGGGGAAGGAATGATTTGACTTTG CGATTGGTTCCCAGTTCCAGAGGTCTCCAGCTATCGGTTTGATCATATCGTTCAAGCGTTCGTAGTCGTTCGTCG TGTTCGCTTTCATTATCCTCGTCATGTTTTCTGGTCTTGTTTTGTATCCTGTAGTTTGATTAGGTAGCAGCCTTG ATGTCGTGCCAATAAGAAATTCGCTTTTTGTTACAGA |
| Length | 2887 |
Gene
| Sequence id | CopciAB_441896.T0 |
|---|---|
| Sequence |
>CopciAB_441896.T0 GTTCTCTTCTTCTTGCATTTACCACTTCGACGCGAACGTCCACTTCTGCCTTGGATCTACGTTGCGCGAAACCTT GCTCTCGACCAAGGTCGCAGAGTACCCGTCTCGCCATTTCACTGGCTCTTCAGCGATACCTCTTCTCATCCACAA CCCCGCCTAGCGACGATGGCTTCCGACATGTATCGGTCCTCGTTCTGCGACTTTAGCGATTCATCTGAGGTGTGA TACGCTCATGCAAACTCTTATTATTGAAGGTGACGAACCCGCCTTTGGACAGTGCACCGATACCTCGACGGGAAC CGTTCTTCGTGATCTCTCCCTCTCACCTCGACCTTCGGTTGTCGACGGACTTGGGGAAGTCTCTACCAACAGTAC AACCCCTCCCCGCCTTGACGTGGAATGGCCCAATTCGCCTCCGTTGGCGGTGAGCTTCATTCCCATCCAGTTTCC CTAGAATCGCCTAACTGATTCGTATTTGGCCAGGTTGCCCTCAGGTTCTCGGGCATATTCGACGATGCTGATGTC GCAGAGATCGAAGGAGATCTTACTCGCCAGACTCACCAAAGCAGATCGCCCACTTTCGACATGACCTATGACATT CATGTAAGGTCTAGTTGAAGATGGGCGAGAAGGACGCTAACCCAGCTTCATTAGAGCTTCCCGAATATCAAGGAC ATCAACTTGGAGATGGGGAACGACAGTGACATCTCCGTAAGTCCTTGCGTTTCTGCAGCTGGAGTTGCTTAAGTT AATCGTGAGGCCGGCGGTGGGTAGGAGGTTGTGGTCATGTCGCAGAAACAGCTCCCCACCGCGCTTTCTACGATC CCAGAGGCAAATGAGCCAAGCCCTGTTCTTGCGGTCCGTGGTCTTCTTCATTATCTGTTGCCTGCTCACTGATTC GTGTTAATCATGTCCAGTATACGACGAGCCTGTCCGACTTTGATATCATCTCCTCTTACAATGCCCCTGATTCTG ATACTCGTGTTATGATCTGCCAAAGGAAAAGGAATGGAAACCTTTACAAGATTCGTTCTCGCCGCATCTATGACC ATTTTCCCTGGATGGAGCAAGACATCCTGGAGACGCTACGAGATATGGGAGCTCCCTTCCTGCCCTATCTGAAGT CTGTCTTCCGAGAGGAAAATCAAGTGCATCTTGTTCTGGTGAGTTCTTCTGTGTACTCTTTACGCGCATACCCCT GACGACTGTGTTACCTGAAGGATTATTGGCCTTCAGCGAGTTTGAGAGAGCTCCTCGCAAAGTACGGAACTCTTG GCTCTCAGCGTGTCCTGTTCTATGCTTCTGAACTCGTTCGTCTTCTCCGCTCACTGCGTGGTTTCCGCTCACCTG CTCTCTTAGCTGACTGGGATTGCCAACCTCCACGGTGTCGGGATCATGCATCGCAACCTTAACCCTGACGATATC ATCTTGGACAAGCGGGGGCACATTGTCATTACCAACTTTGAATTCGCCAACACGATTCCCCCTTCCGGTGTTAAC GCCAATTCGTTTAACAGTGATTTGGTTTTGTCGATACCGAGGTTCCATGGATATCAGGCCCCAGAGATGGTTCTG GGTTGGGCGCATGATATCTCCGTTGATTGCTGGGGATTTGGGATGTTGCTTTACTTCATGTACTTTGGGAAGGTG CGCCACGTTTTCTGCGATCTTGGGACTTGTCGTCCTAAATCATCTGTAGCACCCCTTCCAAGAGGACAGAAACCC GGTAGATCATCGCGTCTTGCGCGCACGCATCATTCAAGGCTCTTTGTCTCCAGAGTCTCTCAGGTTGATTCATCC GCACACGAGGGATATCATCTTGAAGGTACGTTGCTCTTCATCCGCCTGTGTTATGGGAGTAATCAGTTTTTCCTA GTGTGTCGAAAGGAATCCGCGTATTCGATTGAATGTCGATGGGATTAAGAACCACGAATATTTTAACAACGTGTA AGTCACGCTCCACTTGCCCAGTTGACCTGAAACTGACACAATGCTTCAGGCCTTGGGGAAAGGTGGCCGATAAGC TCGTCGAAGGTCAGTTAAACGAGTATTTATTTCAGAGACTAAAACTCATTATACCCGACCACATGTAGTCCCACC ACTACCCGGGTTCTTCAACGATAATGGATCCAGTGGGGAGCTTTTGAGAAGGTCCCATTCGTCAGGCTTGCGACA CACCCTCTACCAAGAGGGAGCCGAGCCTGTTGTCGCAACACCTGAGCAACCTCGACCTGCGCTACTGAGTCCTAT CCCTTCCCATCGTCCTTCTCACCCGCTTCCTCTCTCCCCTTCTCCTTCCGCTGTCACATTCGCCTCTCGTCCCAT GTCTCCATCTGTCCTGACTGAGATGTCAGAACCTCTGGAGGGCAATCTCAGCATCCAATCTCCTAAGCCCCGGAC GTTTCGACCCTTTCTCGACCCTCAGGCGAGCGCTCAGTCGGCGAAAGGAGGACAAGTAGACTCGGGCATGGAAAT CTGGGATCTACTGGATCGTGAGGAGCAAGGCTCGATCGCCTCTTCCAACCTGGGCCGTAGCCACGTTCTTGGATT CCTCAGGCCTCGTAAGCTTCGCAAGAACAATTCGTCGACCAGTCTCTTCAGGCCGTTCGTACTCAACCTGAGCAC GTCGTCCTTGACTCGTGGGTTGAAGAAGAAGAAGTCTAGAAGCTCCACTCGCTTGGGCGAGAGTTCCGCTACCGT ACATGACCCTCAGCAGCGCAACCAGGACGTTGAGCTCCCAGCCGGAATCGAACATATCGGTAGTGGAATTGGATT CCACCGTAATACTCTCTCCTCTGATTCAGGCCATTCGAGGTCTTCAGCTTCCTCAAACTTGCCCAGGTTCTGCCG CGGAGGTGTCAAGCTTGGTCATCGATTCTCATTCAGTGCTTTGCGGCCCAGCGGTGCTCGCAAGATCTTTGGAAG GAATACCCGCTCGAGGTCTCTCTCTTCTACTATGGCGACCAATTCCGGGGATGCTTCTGCACTCGGGGGCCTCCG GTTGGCTCACAATGGGAATAGCCCTGCCTATGTTCACGGTCACACAGGAGCTCTTCCCGGTTCACATACTTCTCC TAGGACGCCTCACGAGGTGCAAGCTAGTACTCCTATCCTCCAAATGAATCCCATGTCACCTAGCGGTTCGTCATC CACCTCCGCTTCTGCGTCATCATCTCCTAGGACCGAGGATGGCCCATTGACTCCAACCACTGTGGCTTGCGACGA GGAGTGTCAAGCCCGAGTATCCTCCAAGGCAGTGATGGACTTTGACGAGGTCAACAAAGAAAGGGGAAGGAATGA TTTGACTTTGCGATTGGTTCCCAGTTCCAGAGGTCTCCAGCTATCGGTTTGATCATATCGTTCAAGCGTTCGTAG TCGTTCGTCGTGTTCGCTTTCATTATCCTCGTCATGTTTTCTGGTCTTGTTTTGTATCCTGTAGTTTGATTAGGT AGCAGCCTTGATGTCGTGCCAATAAGAAATTCGCTTTTTGTTACAGA |
| Length | 3497 |