CopciAB_444565
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_444565 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 444565 |
| Uniprot id | Functional description | Serine/Threonine protein kinases, catalytic domain | |
| Location | scaffold_5:1777666..1780587 | Strand | - |
| Gene length (nt) | 2922 | Transcript length (nt) | 2463 |
| CDS length (nt) | 1968 | Protein length (aa) | 655 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7259762 | 76.1 | 0 | 1039 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1453572 | 73.8 | 0 | 1007 |
| Pleurotus ostreatus PC9 | PleosPC9_1_49007 | 73.9 | 0 | 1004 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB22823 | 72.4 | 0 | 1001 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1092749 | 73.9 | 4.941E-304 | 991 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_219768 | 73.2 | 1.481E-304 | 981 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_73338 | 72.6 | 8.295E-304 | 976 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_14262 | 73.8 | 6.263E-304 | 968 |
| Lentinula edodes NBRC 111202 | Lenedo1_1034727 | 73.8 | 1.4E-304 | 967 |
| Lentinula edodes B17 | Lened_B_1_1_10206 | 73.9 | 1.717E-304 | 952 |
| Schizophyllum commune H4-8 | Schco3_2621466 | 72.5 | 8.459E-304 | 936 |
| Flammulina velutipes | Flave_chr11AA00599 | 69.5 | 9.754E-301 | 924 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_41754 | 66.1 | 2.749E-288 | 888 |
| Grifola frondosa | Grifr_OBZ76590 | 65.9 | 9.165E-274 | 846 |
| Auricularia subglabra | Aurde3_1_1272538 | 61.3 | 3.909E-272 | 842 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_444565.T0 |
| Description | Serine/Threonine protein kinases, catalytic domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd06609 | STKc_MST3_like | - | 34 | 304 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 36 | 284 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
| IPR017441 | Protein kinase, ATP binding site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K08286 |
| K08838 |
| K21618 |
EggNOG
| COG category | Description |
|---|---|
| T | Serine/Threonine protein kinases, catalytic domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
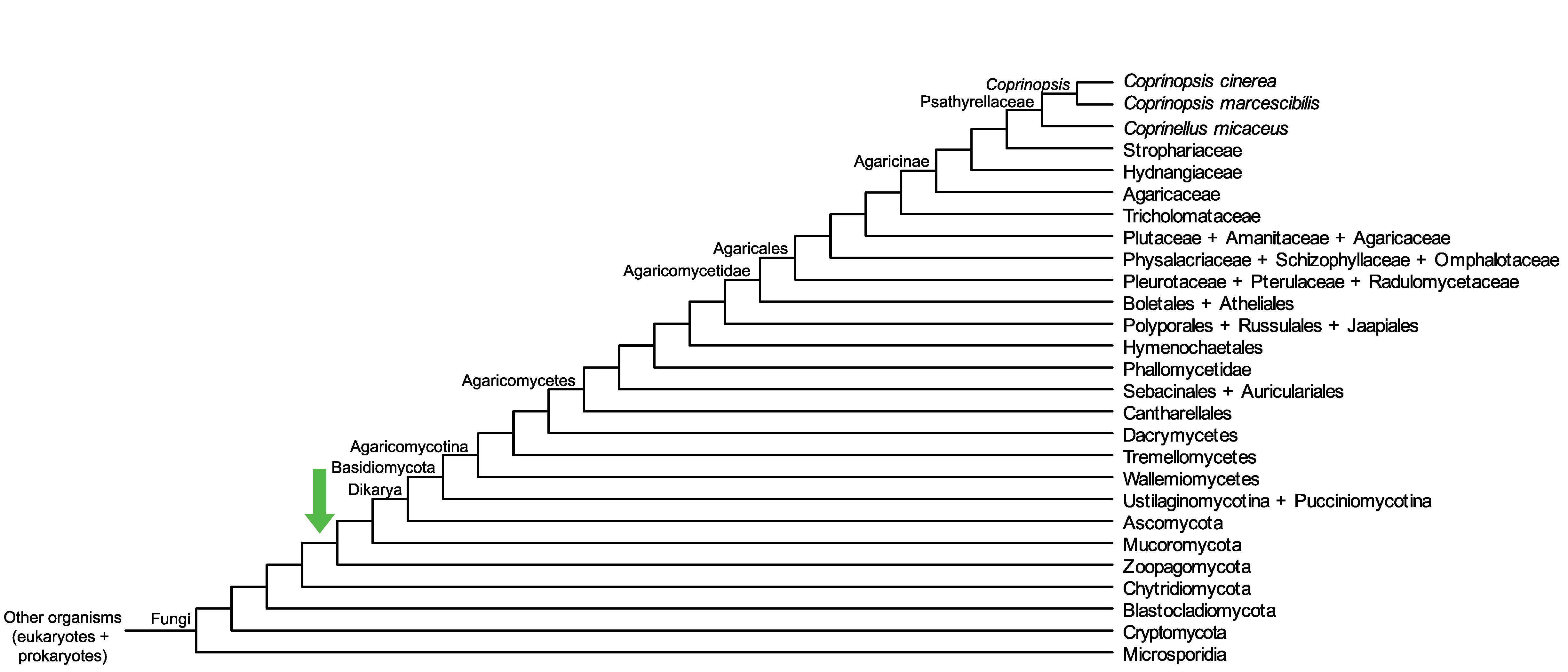
Conservation of CopciAB_444565 across fungi.
Arrow shows the origin of gene family containing CopciAB_444565.
Protein
| Sequence id | CopciAB_444565.T0 |
|---|---|
| Sequence |
>CopciAB_444565.T0 MTKSSTRSPPPSPKVEQRTRPRDPCPIPSSNPASQYTLLEKLGTGSFGVVYKAIHNETKQIVAIKQIDLEDSDDD ISEIQQEIASLAQCDSEYVTKYYGSFVVHYKLWIVMEYLAGGSCLDLLKAGPFSEAHIAVICRELLLGLDYLHAE GTIHRDIKAANVLLSSSGKVKLADFGVAAQLTNTLRHTFVGTPFWMAPEVIRQAGYDSKADMWSLGITAIEMAKG EPPLAEYHPMRVLFLIPKAKPPTLEGPFSAAFKDFVTLCLTKDPKLRPSANELLQHRFIRGAKKTSYLTELIERY QEWRMRSPHKGQNNAATIRNTATWDMNDTIRSDWNFDTVKSMGAMGTFRGSVKDLGMPTGMILEADESDDGETSI DTGAATKGSDPLLSNSQAAHSTVMIKNPPPHLAESDDIADDDIPNGAPPAYSGSVRSARRASYAERSSTDGPGTV LNEADLGTGVDTIRPVKKVDAAGSLRLSTGYLGSIRKEGSGSSYSTHKRSASEGARAGRAMVDEVVLPVLQRAIR DDMDAREIESLSMLQRGFEELKEANSELAYNVILDILQGINDNNAIKRHVQTSRGLFPHKRITRKSEMTSKGLVV TEETEEVSGLPESSNGVPPAQAEAETPRRSPIAELLYMRWLEGLRLKWPSIMTGS |
| Length | 655 |
Coding
| Sequence id | CopciAB_444565.T0 |
|---|---|
| Sequence |
>CopciAB_444565.T0 ATGACAAAGTCTTCGACCCGCTCGCCGCCCCCCTCTCCCAAGGTGGAGCAACGGACCCGACCCAGGGATCCCTGC CCAATCCCGAGCAGCAACCCCGCAAGCCAGTATACGCTACTGGAGAAGCTTGGGACAGGAAGTTTTGGTGTCGTG TACAAGGCCATACACAACGAGACCAAGCAAATTGTTGCAATCAAGCAGATAGATCTTGAAGACTCGGACGACGAC ATTTCTGAAATACAGCAAGAAATTGCCAGCCTTGCCCAATGCGACTCTGAATACGTGACCAAGTACTATGGGTCC TTCGTGGTGCACTACAAACTCTGGATCGTGATGGAGTACCTCGCAGGTGGCTCGTGTCTTGACCTTCTTAAAGCC GGGCCGTTTTCTGAAGCACACATCGCTGTCATTTGCCGCGAGCTTCTTTTGGGACTCGACTACCTCCACGCTGAA GGAACCATTCACCGGGACATCAAGGCTGCGAATGTGCTTCTCTCCTCGTCGGGCAAAGTCAAGCTGGCCGATTTT GGCGTTGCAGCTCAGCTCACCAACACCCTACGACACACATTTGTCGGCACCCCCTTCTGGATGGCACCTGAAGTT ATTCGACAGGCCGGATACGATTCCAAAGCGGACATGTGGAGCCTTGGAATTACAGCGATTGAGATGGCTAAAGGC GAGCCTCCATTAGCCGAGTATCACCCGATGCGGGTTCTTTTTCTCATCCCAAAAGCGAAGCCACCGACTCTTGAA GGACCGTTCTCAGCGGCATTCAAGGATTTCGTGACCCTATGCTTGACTAAAGACCCCAAACTCCGCCCATCTGCC AACGAGCTGCTTCAACATAGATTCATTCGTGGCGCAAAGAAGACATCTTATTTGACAGAGTTGATCGAGCGGTAC CAGGAATGGCGCATGCGGAGCCCTCACAAAGGCCAGAATAATGCTGCTACGATCAGGAATACCGCCACATGGGAT ATGAATGATACCATTCGGAGCGACTGGAATTTCGACACTGTGAAGTCGATGGGGGCCATGGGTACTTTCAGGGGC TCGGTTAAGGACCTTGGGATGCCTACGGGGATGATCCTTGAAGCCGACGAGAGCGACGATGGAGAGACCTCGATT GATACAGGTGCAGCAACCAAGGGAAGCGATCCGCTTCTCTCAAATTCACAAGCCGCACATTCGACTGTGATGATC AAGAACCCTCCGCCGCATCTTGCGGAATCAGACGACATTGCAGACGATGATATCCCAAACGGTGCCCCTCCTGCC TATTCCGGTTCGGTTCGCAGTGCCAGACGCGCCTCTTACGCAGAGAGATCCTCCACAGATGGTCCAGGAACGGTT TTAAATGAGGCAGATCTTGGTACTGGAGTGGATACTATCCGACCCGTCAAGAAAGTCGATGCAGCTGGCTCTCTG CGCCTGTCCACTGGTTATCTCGGTAGCATACGCAAGGAGGGTAGCGGCTCATCCTACTCGACTCACAAACGGTCG GCAAGTGAAGGTGCCAGGGCCGGTAGGGCAATGGTAGACGAGGTTGTATTGCCTGTCCTACAAAGAGCTATTCGG GATGACATGGACGCACGCGAAATCGAGTCTCTCAGCATGCTTCAGCGAGGCTTCGAAGAACTCAAGGAAGCCAAC TCGGAACTGGCGTACAACGTCATTCTGGACATCCTCCAGGGCATCAATGATAACAACGCCATTAAACGACATGTC CAGACATCCCGCGGGCTGTTCCCACACAAACGCATTACTCGCAAGAGTGAGATGACGTCCAAGGGTCTCGTCGTC ACAGAAGAGACGGAGGAGGTATCCGGACTTCCGGAGTCGTCGAACGGTGTTCCTCCTGCGCAGGCTGAAGCTGAA ACCCCGCGACGGTCCCCCATAGCTGAACTCCTCTACATGCGCTGGCTGGAAGGACTGAGATTGAAGTGGCCTAGT ATCATGACCGGCAGCTAG |
| Length | 1968 |
Transcript
| Sequence id | CopciAB_444565.T0 |
|---|---|
| Sequence |
>CopciAB_444565.T0 GACTCTTCCTTCCACCACCACCGTCGTCCCCGTCCTACACACAAGCCGAGTCTCTGTCGTGCCCTCATCGCCAAC GACGGTCCCCGACGAATCCCCCGCCTTTCCCAACCCTCAACGTCCGCGACACCCCAATTCGAGAGACTTGGCTTT TTCTCTTTCTCTCAACTTTCTCTCTCTTTTTTCCGCTTTTTTCACTTGTATTCCTGTTAACGACTTCTTAACGAC CTTATCGACGATCTAACGACATTTACGAATACGATTATGACAAAGTCTTCGACCCGCTCGCCGCCCCCCTCTCCC AAGGTGGAGCAACGGACCCGACCCAGGGATCCCTGCCCAATCCCGAGCAGCAACCCCGCAAGCCAGTATACGCTA CTGGAGAAGCTTGGGACAGGAAGTTTTGGTGTCGTGTACAAGGCCATACACAACGAGACCAAGCAAATTGTTGCA ATCAAGCAGATAGATCTTGAAGACTCGGACGACGACATTTCTGAAATACAGCAAGAAATTGCCAGCCTTGCCCAA TGCGACTCTGAATACGTGACCAAGTACTATGGGTCCTTCGTGGTGCACTACAAACTCTGGATCGTGATGGAGTAC CTCGCAGGTGGCTCGTGTCTTGACCTTCTTAAAGCCGGGCCGTTTTCTGAAGCACACATCGCTGTCATTTGCCGC GAGCTTCTTTTGGGACTCGACTACCTCCACGCTGAAGGAACCATTCACCGGGACATCAAGGCTGCGAATGTGCTT CTCTCCTCGTCGGGCAAAGTCAAGCTGGCCGATTTTGGCGTTGCAGCTCAGCTCACCAACACCCTACGACACACA TTTGTCGGCACCCCCTTCTGGATGGCACCTGAAGTTATTCGACAGGCCGGATACGATTCCAAAGCGGACATGTGG AGCCTTGGAATTACAGCGATTGAGATGGCTAAAGGCGAGCCTCCATTAGCCGAGTATCACCCGATGCGGGTTCTT TTTCTCATCCCAAAAGCGAAGCCACCGACTCTTGAAGGACCGTTCTCAGCGGCATTCAAGGATTTCGTGACCCTA TGCTTGACTAAAGACCCCAAACTCCGCCCATCTGCCAACGAGCTGCTTCAACATAGATTCATTCGTGGCGCAAAG AAGACATCTTATTTGACAGAGTTGATCGAGCGGTACCAGGAATGGCGCATGCGGAGCCCTCACAAAGGCCAGAAT AATGCTGCTACGATCAGGAATACCGCCACATGGGATATGAATGATACCATTCGGAGCGACTGGAATTTCGACACT GTGAAGTCGATGGGGGCCATGGGTACTTTCAGGGGCTCGGTTAAGGACCTTGGGATGCCTACGGGGATGATCCTT GAAGCCGACGAGAGCGACGATGGAGAGACCTCGATTGATACAGGTGCAGCAACCAAGGGAAGCGATCCGCTTCTC TCAAATTCACAAGCCGCACATTCGACTGTGATGATCAAGAACCCTCCGCCGCATCTTGCGGAATCAGACGACATT GCAGACGATGATATCCCAAACGGTGCCCCTCCTGCCTATTCCGGTTCGGTTCGCAGTGCCAGACGCGCCTCTTAC GCAGAGAGATCCTCCACAGATGGTCCAGGAACGGTTTTAAATGAGGCAGATCTTGGTACTGGAGTGGATACTATC CGACCCGTCAAGAAAGTCGATGCAGCTGGCTCTCTGCGCCTGTCCACTGGTTATCTCGGTAGCATACGCAAGGAG GGTAGCGGCTCATCCTACTCGACTCACAAACGGTCGGCAAGTGAAGGTGCCAGGGCCGGTAGGGCAATGGTAGAC GAGGTTGTATTGCCTGTCCTACAAAGAGCTATTCGGGATGACATGGACGCACGCGAAATCGAGTCTCTCAGCATG CTTCAGCGAGGCTTCGAAGAACTCAAGGAAGCCAACTCGGAACTGGCGTACAACGTCATTCTGGACATCCTCCAG GGCATCAATGATAACAACGCCATTAAACGACATGTCCAGACATCCCGCGGGCTGTTCCCACACAAACGCATTACT CGCAAGAGTGAGATGACGTCCAAGGGTCTCGTCGTCACAGAAGAGACGGAGGAGGTATCCGGACTTCCGGAGTCG TCGAACGGTGTTCCTCCTGCGCAGGCTGAAGCTGAAACCCCGCGACGGTCCCCCATAGCTGAACTCCTCTACATG CGCTGGCTGGAAGGACTGAGATTGAAGTGGCCTAGTATCATGACCGGCAGCTAGGCAGCTGGCGCCATGTCCTTT GGCACAATGTACAACTCTTGTATATACCCTTCTTCCTCTTGGTTATTTATGTACTGTAGGTTGTTTCACGATCAT TCGCTAATGCTCCCTCGCTTCTACCACTATCTCCGCTTGCAACTTTTGGCTATTCCTTGGACGTAACGTTGAATT AAGCTGTACGATGTACATATCTGCTTGTAGTACCATTTAACGAATTCTGATGACGGTTACTTG |
| Length | 2463 |
Gene
| Sequence id | CopciAB_444565.T0 |
|---|---|
| Sequence |
>CopciAB_444565.T0 GACTCTTCCTTCCACCACCACCGTCGTCCCCGTCCTACACACAAGCCGAGTCTCTGTCGTGCCCTCATCGCCAAC GACGGTCCCCGACGAATCCCCCGCCTTTCCCAACCCTCAACGTCCGCGACACCCCAATTCGAGAGACTTGGCTTT TTCTCTTTCTCTCAACTTTCTCTCTCTTTTTTCCGCTTTTTTCACTTGTATTCCTGTTAACGACTTCTTAACGAC CTTATCGACGATCTAACGACATTTACGAATACGATTATGGTGAGTTTCGATCTCATAATCGTCGAGAAGATCCCT TCGCTCTGGCCCCGCACGTGTGGGCACGAAGCTGTGAATAATGCATGAGGTCACCCTGAGAGCGCCTCGTCGGCG AATTTCTTTAAATCCTTCTTCAATACCTTTCTCTTTTTCTTTTCGACATGACACTAACTTGGGTATGTTTCGTAA ACAGACAAAGTCTTCGACCCGCTCGCCGCCCCCCTCTCCCAAGGTGGAGCAACGGACCCGACCCAGGGATCCCTG CCCAATCCCGAGCAGCAACCCCGCAAGCCAGTATACGCTACTGGAGAAGCTTGGGACAGGAAGTTTTGGTGTCGT GTACAAGGCCATACACAACGAGACCAAGCAAATTGTTGCAATCAAGCAGATAGGTGCGCCCTCCTCGTTGTCTTG GCTCCGCACGGTACTAATCCAATATGGTAGATCTTGAAGACTCGGACGACGACATTTCTGAAATACAGCAAGAAA TTGCCAGCCTTGCCCAATGCGACTCTGAATACGTGACCAAGTACTATGGGTCCTTCGTGGTGCACTACAAACTCT GGATCGTGATGGAGTACCTCGCAGGTGGCTCGTGTCTTGACCTTCTTAAAGCCGGGCCGTTTTCTGAAGCACACA TCGCTGTCATTTGCCGCGAGCTTCTTTTGGGACTCGACTACCTCCACGCTGAAGGAACCATTCACCGGGACATCA AGGCTGCGAATGTGCTTCTCTCCTCGTCGGGCAAAGTCAAGCTGGCCGATTTTGGCGTTGCAGCTCAGCTCACCA ACACCCTACGACACACATTTGTCGGCACCCCCTTCTGGATGGCACCTGAAGTTATTCGACAGGCCGGATACGATT CCAAAGCGGACATGTGGAGCCTTGGAATTACAGCGATTGAGATGGCTAAAGGCGAGCCTCCATTAGCCGAGTATC ACCCGATGCGGGTTCTTTTTCTCATCCCAAAAGCGAAGCCACCGACTCTTGAAGGACCGTTCTCAGCGGCATTCA AGGATTTCGTGACCCTATGCTTGACTAAAGACCCCAAACTCGTTCGTAATTCTAATCTACCCTGTTTGAAATACA TTACTAACTCCTTATTGTGTTCAATTAGCGCCCATCTGCCAACGAGCTGCTTCAACATAGATTCATTCGTGGCGC AAAGAAGACATCTTATTTGACAGAGTTGATCGAGCGGTACCAGGAATGGCGCATGCGGAGCCCTCACAAAGGCCA GAATAATGCTGCTACGATCAGGAATACCGCCACATGGGATATGAATGATACCATTCGGAGCGACTGGAATTTCGA CACTGTGAAGTCGATGGGGGCCATGGGTACTTTCAGGGGCTCGGTTAAGGACCTTGGGATGCCTACGGGGATGAT CCTTGAAGCCGACGAGAGCGACGATGGAGAGACCTCGATTGATACAGGTGCAGCAACCAAGGGAAGCGATCCGCT TCTCTCAAATTCACAAGCCGCACATTCGACTGTGATGATCAAGAACCCTCCGCCGCATCTTGCGGAATCAGACGA CATTGCAGACGATGATATCCCAAACGGTATGCATCTAACGTCTGGCTCGGTGTTAACTTTACTGACCATCCTCTT TAGGTGCCCCTCCTGCCTATTCCGGTTCGGTTCGCAGTGCCAGACGCGCCTCTTACGCAGAGAGATCCTCCACAG ATGGTCCAGGAACGGTTTTAAATGAGGCAGATCTTGGTACTGGAGTGGATACTATCCGACCCGTCAAGAAAGTCG ATGCAGCTGGCTCTCTGCGCCTGTCCACTGGTTATCTCGGTAGCATACGCAAGGAGGGTAGCGGCTCATCCTACT CGACTCACAAACGGTCGGCAAGTGAAGGTGCCAGGGCCGGTAGGGCAATGGTAGACGAGGTTGTATTGCCTGTCC TACAAAGAGTAAGTGCCTTGCGGCTACGCCGTCATGAGATTTGCTCAAGTGTTTTTCAGGCTATTCGGGATGACA TGGACGCACGCGAAATCGAGTCTCTCAGCATGCTTCAGCGAGGCTTCGAAGAACTCAAGGAAGCCAACTCGGAAC TGGCGTACAACGTCATTCTGGACATCCTCCAGGGCATCAATGAGTGAGTAAATCAACCACTCTCATGGAACGCCA GTCCCTAACTATGTTTTCAGTAACAACGCCATTAAACGACATGTCCAGACATCCCGCGGGCTGTTCCCACACAAA CGCATTACTCGCAAGAGTGAGATGACGTCCAAGGGTCTCGTCGTCACAGAAGAGACGGAGGAGGTATCCGGACTT CCGGAGTCGTCGAACGGTGTTCCTCCTGCGCAGGCTGAAGCTGAAACCCCGCGACGGTCCCCCATAGCTGAACTC CTCTACATGCGCTGGCTGGAAGGACTGAGATTGAAGTGGCCTAGTATCATGACCGGCAGCTAGGCAGCTGGCGCC ATGTCCTTTGGCACAATGTACAACTCTTGTATATACCCTTCTTCCTCTTGGTTATTTATGTACTGTAGGTTGTTT CACGATCATTCGCTAATGCTCCCTCGCTTCTACCACTATCTCCGCTTGCAACTTTTGGCTATTCCTTGGACGTAA CGTTGAATTAAGCTGTACGATGTACATATCTGCTTGTAGTACCATTTAACGAATTCTGATGACGGTTACTTG |
| Length | 2922 |