CopciAB_445701
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_445701 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | EXG3 | Synonyms | 445701 |
| Uniprot id | Functional description | Belongs to the glycosyl hydrolase 5 (cellulase A) family | |
| Location | scaffold_3:42625..45738 | Strand | - |
| Gene length (nt) | 3114 | Transcript length (nt) | 2392 |
| CDS length (nt) | 1683 | Protein length (aa) | 560 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Saccharomyces cerevisiae | YBR056W_YBR056W |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7270598 | 50.4 | 7.417E-163 | 520 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB15978 | 50.8 | 6.011E-157 | 503 |
| Schizophyllum commune H4-8 | Schco3_2495155 | 46.2 | 2.728E-146 | 472 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_69695 | 45.9 | 6.36E-140 | 453 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_205367 | 45.5 | 1.448E-138 | 449 |
| Flammulina velutipes | Flave_chr02AA00152 | 46.1 | 1.346E-131 | 429 |
| Grifola frondosa | Grifr_OBZ65555 | 44.2 | 1.537E-129 | 423 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_57596 | 54.1 | 1.05E-123 | 406 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_15094 | 36.8 | 2.476E-97 | 329 |
| Lentinula edodes NBRC 111202 | Lenedo1_1164141 | 36.4 | 2.628E-96 | 326 |
| Lentinula edodes B17 | Lened_B_1_1_13392 | 35.6 | 2.193E-76 | 267 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | EXG3 |
|---|---|
| Protein id | CopciAB_445701.T0 |
| Description | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 118 | 405 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR017853 | Glycoside hydrolase superfamily |
| IPR001547 | Glycoside hydrolase, family 5 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| K01210 |
EggNOG
| COG category | Description |
|---|---|
| G | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH5 | GH5_50 |
Transcription factor
| Group |
|---|
| No records |
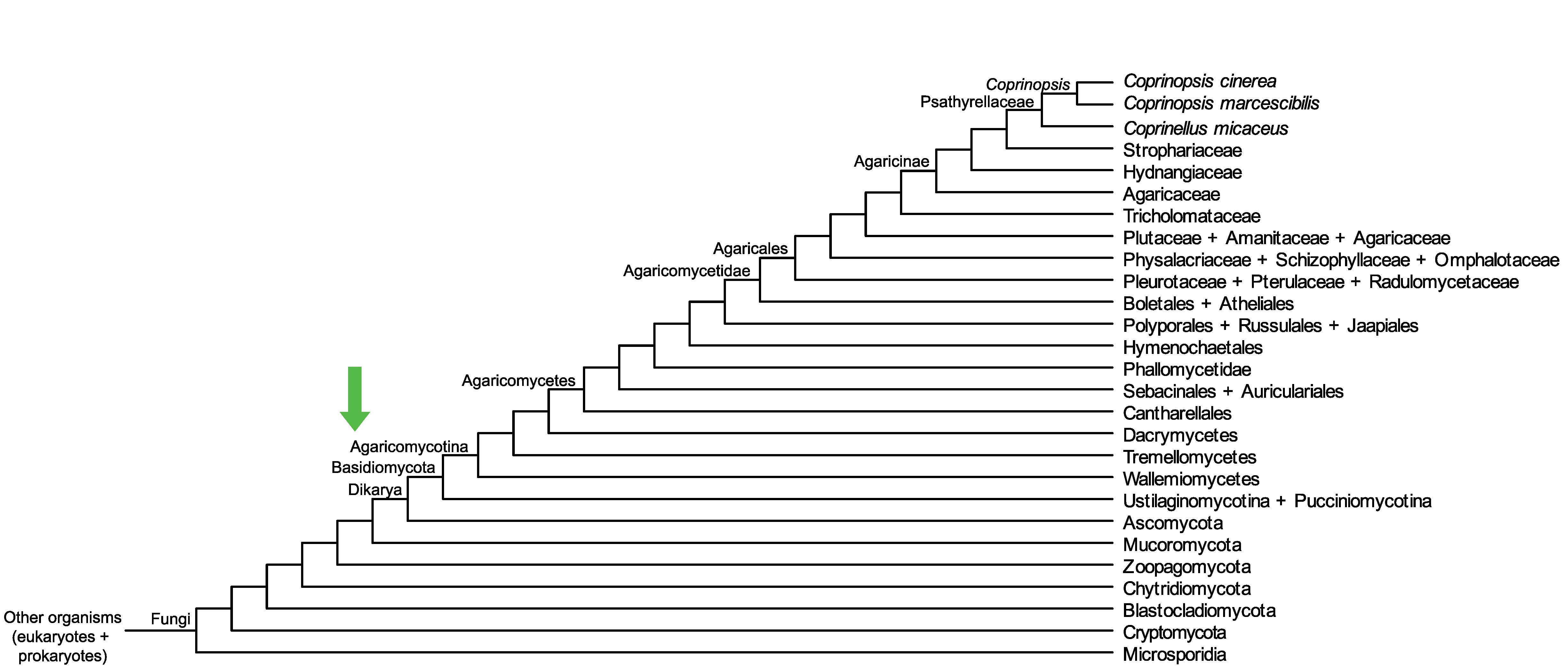
Conservation of CopciAB_445701 across fungi.
Arrow shows the origin of gene family containing CopciAB_445701.
Protein
| Sequence id | CopciAB_445701.T0 |
|---|---|
| Sequence |
>CopciAB_445701.T0 MRRFFQGFKSDVDKFKDQVQEHVAQFSPGDVSSPGVEDFDEHESEHTKIFPVRQEDLLKYRKHRGVNLGSWFVLE RWITDSPFREAAAPAASDLDVAKGRNAKSILERHWDTWIQPADWDWIAARGINSVRIPIGYYHVCGADRSILDGT DFWPFYDVYQGAWRRITNAILEANKRGITVQLDMHAAPGKQNADPHSGTSNPANFFHDPHNLRRGLYALQSLSMH LTSFLNSHDPPLSNVVSIELVNEPAPPNDGVLKRWYDDAIRVVQRGARGVPVYIGECWRPEVYAEYVARELKQED PEGLVVLDHHLYRCFTQEDINTSVYDHTGRLRDHNGILSTFTRVAELVGRAGGALSVCEWSGGLNEGSLKHVRGE DQRRNARRDFLRAQLELYERTCAGWYFWTYKKEYRGDVGWSFREAVEEGVFPMKNGRIDAGILVRKKRWTDADVE RECVRRVGKMEEERKKAYDAHRSYWSRIPGRYEHRRFWYGFTAGWNMAWRFYDSIEPDGIGNPVSEPGPLHALAR RSSVYPVEGPHYWEYEHGFLQGVGSARGDFEGEFC |
| Length | 560 |
Coding
| Sequence id | CopciAB_445701.T0 |
|---|---|
| Sequence |
>CopciAB_445701.T0 ATGCGCCGATTTTTCCAAGGATTCAAGAGCGACGTTGATAAATTCAAGGACCAAGTGCAAGAACACGTTGCCCAG TTCAGTCCAGGAGATGTATCCTCCCCCGGGGTAGAAGATTTCGACGAGCACGAATCGGAACACACCAAGATCTTC CCCGTCCGACAAGAGGATTTGTTGAAGTACCGGAAACATCGTGGAGTAAACCTCGGCTCGTGGTTCGTCCTCGAA CGCTGGATAACAGATTCACCCTTCCGCGAAGCAGCAGCACCCGCAGCGAGCGATCTGGACGTCGCGAAAGGCCGA AACGCCAAATCCATTTTGGAACGACATTGGGATACGTGGATCCAACCGGCGGACTGGGACTGGATTGCTGCGAGG GGTATTAATTCTGTCCGGATTCCTATTGGGTATTATCATGTATGCGGAGCGGATAGGAGTATACTCGACGGGACG GACTTTTGGCCGTTTTATGATGTGTACCAGGGCGCGTGGAGGAGGATTACGAATGCCATCCTCGAGGCGAACAAG CGCGGTATAACCGTCCAACTCGACATGCACGCAGCCCCAGGAAAACAAAACGCCGACCCGCACTCCGGCACCTCC AACCCCGCCAACTTCTTCCACGACCCGCACAACCTCCGTCGGGGCCTCTACGCTCTGCAGTCCCTCTCCATGCAC CTCACGTCTTTCCTCAACAGCCACGACCCGCCCCTCTCCAACGTCGTCTCCATCGAATTGGTCAACGAACCTGCA CCGCCTAACGATGGTGTGCTGAAGCGGTGGTACGACGATGCGATAAGGGTTGTGCAGAGGGGTGCGCGTGGCGTG CCGGTATATATTGGGGAGTGTTGGAGGCCGGAGGTGTATGCTGAGTATGTGGCGAGGGAGCTGAAGCAGGAGGAT CCAGAAGGCCTAGTAGTACTAGACCACCACCTCTACCGTTGCTTCACCCAAGAAGACATCAACACATCCGTCTAC GACCACACCGGGCGGCTGCGCGACCACAACGGCATACTCTCGACCTTTACGCGCGTCGCTGAGCTCGTGGGTCGA GCCGGCGGGGCGCTATCCGTGTGCGAGTGGTCTGGCGGTCTCAATGAGGGGAGTTTGAAGCATGTGCGTGGGGAG GACCAACGGAGGAATGCGAGACGTGATTTTTTGAGAGCGCAGTTGGAGTTGTATGAGCGGACGTGTGCGGGGTGG TATTTTTGGACGTATAAGAAGGAGTATAGGGGTGATGTGGGGTGGTCGTTTAGGGAGGCGGTGGAGGAGGGGGTG TTTCCTATGAAGAATGGACGAATTGACGCTGGTATTTTGGTGAGGAAGAAGAGGTGGACGGATGCGGATGTGGAA AGGGAATGTGTGCGTAGGGTGGGGAAGATGGAGGAGGAGAGGAAGAAGGCGTATGATGCACATAGATCCTACTGG TCCCGCATCCCGGGGCGCTACGAGCACCGCAGGTTCTGGTATGGTTTTACAGCTGGGTGGAATATGGCGTGGAGG TTTTATGATTCTATCGAACCGGATGGCATAGGCAACCCAGTAAGCGAACCAGGCCCGCTACACGCCCTCGCGCGG AGAAGTTCGGTGTACCCCGTTGAGGGCCCGCATTATTGGGAGTATGAGCATGGGTTTTTGCAGGGTGTGGGGAGT GCTAGAGGGGATTTTGAGGGGGAGTTTTGCTAA |
| Length | 1683 |
Transcript
| Sequence id | CopciAB_445701.T0 |
|---|---|
| Sequence |
>CopciAB_445701.T0 ACGACCATGCAAGCCAAGGTCGCAGTTCACGTTCACAAGTTTGAGACAGATCTTTTGTTTTTTCTTTATCGCCCT TATCGTCCACTGTTGTTCGTTGAACATTGAACGTTGAACGCGGCGACGAGGGCTTCTTGTGTACCTCGAACATCT CTACGTCGGGCTTCTTGTTTTTCGACATCTCTCCTTGATATACGCCTTCTTCAACACATCCTCGTTCCAGGGTCA CCTCAACTCACTTCGGAGCGCCAACCACTATCCCTGCGACCAAATTCAACGAGATGCGCCGATTTTTCCAAGGAT TCAAGAGCGACGTTGATAAATTCAAGGACCAAGTGCAAGAACACGTTGCCCAGTTCAGTCCAGGAGATGTATCCT CCCCCGGGGTAGAAGATTTCGACGAGCACGAATCGGAACACACCAAGATCTTCCCCGTCCGACAAGAGGATTTGT TGAAGTACCGGAAACATCGTGGAGTAAACCTCGGCTCGTGGTTCGTCCTCGAACGCTGGATAACAGATTCACCCT TCCGCGAAGCAGCAGCACCCGCAGCGAGCGATCTGGACGTCGCGAAAGGCCGAAACGCCAAATCCATTTTGGAAC GACATTGGGATACGTGGATCCAACCGGCGGACTGGGACTGGATTGCTGCGAGGGGTATTAATTCTGTCCGGATTC CTATTGGGTATTATCATGTATGCGGAGCGGATAGGAGTATACTCGACGGGACGGACTTTTGGCCGTTTTATGATG TGTACCAGGGCGCGTGGAGGAGGATTACGAATGCCATCCTCGAGGCGAACAAGCGCGGTATAACCGTCCAACTCG ACATGCACGCAGCCCCAGGAAAACAAAACGCCGACCCGCACTCCGGCACCTCCAACCCCGCCAACTTCTTCCACG ACCCGCACAACCTCCGTCGGGGCCTCTACGCTCTGCAGTCCCTCTCCATGCACCTCACGTCTTTCCTCAACAGCC ACGACCCGCCCCTCTCCAACGTCGTCTCCATCGAATTGGTCAACGAACCTGCACCGCCTAACGATGGTGTGCTGA AGCGGTGGTACGACGATGCGATAAGGGTTGTGCAGAGGGGTGCGCGTGGCGTGCCGGTATATATTGGGGAGTGTT GGAGGCCGGAGGTGTATGCTGAGTATGTGGCGAGGGAGCTGAAGCAGGAGGATCCAGAAGGCCTAGTAGTACTAG ACCACCACCTCTACCGTTGCTTCACCCAAGAAGACATCAACACATCCGTCTACGACCACACCGGGCGGCTGCGCG ACCACAACGGCATACTCTCGACCTTTACGCGCGTCGCTGAGCTCGTGGGTCGAGCCGGCGGGGCGCTATCCGTGT GCGAGTGGTCTGGCGGTCTCAATGAGGGGAGTTTGAAGCATGTGCGTGGGGAGGACCAACGGAGGAATGCGAGAC GTGATTTTTTGAGAGCGCAGTTGGAGTTGTATGAGCGGACGTGTGCGGGGTGGTATTTTTGGACGTATAAGAAGG AGTATAGGGGTGATGTGGGGTGGTCGTTTAGGGAGGCGGTGGAGGAGGGGGTGTTTCCTATGAAGAATGGACGAA TTGACGCTGGTATTTTGGTGAGGAAGAAGAGGTGGACGGATGCGGATGTGGAAAGGGAATGTGTGCGTAGGGTGG GGAAGATGGAGGAGGAGAGGAAGAAGGCGTATGATGCACATAGATCCTACTGGTCCCGCATCCCGGGGCGCTACG AGCACCGCAGGTTCTGGTATGGTTTTACAGCTGGGTGGAATATGGCGTGGAGGTTTTATGATTCTATCGAACCGG ATGGCATAGGCAACCCAGTAAGCGAACCAGGCCCGCTACACGCCCTCGCGCGGAGAAGTTCGGTGTACCCCGTTG AGGGCCCGCATTATTGGGAGTATGAGCATGGGTTTTTGCAGGGTGTGGGGAGTGCTAGAGGGGATTTTGAGGGGG AGTTTTGCTAAGGGGTATTCAGGAGAGAGTTTTTGTTGGTAGTGGGCGGGGTGGTGGGGAGGGAGGTTTGTGAGG TTTGTAATTGGGAAGGGGTTTGTTAGCGGGCGGGGTTGGGGAGGGAGGTTTGGTAGCGGGCTGGTTGTTGGGGAG GGGGCGTAGGTTATCTAGGCGTTGTGGGAGGGTACGTTATCTAGGAGTTGTGGGAGGGGAGGAAAGCCAGTAGAA AGAGTGAAATTTGCTTTATTTTTTTGTAGCGTAGCGTAAGCAACAAGTGGAAAGTTGGAGAGTCGCAGTTGCGTC TGGTGGTACTAGTAGGCTACTCTGGGTATATCGTATTATGTATACTAAAGAGCGTGGACGAGGAAAGGGATGATT GTGGACTGGTAGTAGTAAGGTAGATGGAAAAAATGGATCTCCGTTCCTGCAGATGCAGAAGGGTTTG |
| Length | 2392 |
Gene
| Sequence id | CopciAB_445701.T0 |
|---|---|
| Sequence |
>CopciAB_445701.T0 ACGACCATGCAAGCCAAGGTCGCAGTTCACGTTCACAAGTTTGAGACAGATCTTTTGTTTTTTCTTTATCGCCCT TATCGTCCACTGTTGTTCGTTGAACATTGAACGTTGAACGCGGCGACGAGGGCTTCTTGTGTACCTCGAACATCT CTACGTCGGGCTTCTTGTTTTTCGACATCTCTCCTTGATATACGCCTTCTTCAACACATCCTCGTTCCAGGGTCA CCTCAACTCACTTCGGAGCGCCAACCACTATCCCTGCGACCAAATTCAACGAGATGCGCCGATTTTTCCAAGGAT TCAAGAGCGACGTTGATAAATTCAAGGACCAAGTGCAAGAACACGTTGCCCAGTTCAGTCCAGGAGATGTATCCT CCCCCGGGGTAGAAGATTTCGACGAGCACGAATCGGAACACACCAAGATCTTCCCCGTCCGACAAGAGGATTTGT TGAAGTACCGGAAACATCGTGGAGTAAACCTCGGTGCGTCGTCTTGATCGTCTTGCCATGAACATGCACCCAACT CTCCTCATTCTCCTCCCTATGGACGTGCAGGCTCGTGGTTCGTCCTCGAACGCTGGATAACAGATTCACCCTTCC GCGAAGCAGCAGCACCCGCAGCGAGCGATCTGGACGTCGCGAAAGGCCGAAACGCCAAATCCATTTTGGAACGAC ATTGGGATACGTGGATCCAACCGGCGGACTGGGACTGGATTGCTGCGAGGGGTATTAATTCTGTCCGGATTCCTG TACGTGGCTTCTCTTTGTCTGTGGTGACCTCGGTCTGTTCGTATGCTAACTGATGAAATGAGTAGATTGGGTATT ATCATGTATGCGGAGCGGATAGGAGTATACTCGACGGGACGGACTTTTGGCCGTTTTATGATGTGTACCAGGGCG CGTGGAGGAGGATTACGAATGCCATCCTCGAGGCGAACAAGCGCGGTATAACCGTCCAACTCGGTACGTTCCCCA TCCCATCCCATCCCTCCCTCCCTTCCCTCCTGACAAGACTGGACTCCAGACATGCACGCAGCCCCAGGAAAACAA AACGCCGACCCGCACTCCGGCACCTCCAACCCCGCCAACTTCTTCCACGACCCGCACAACCTCCGTCGGGGCCTC TACGCTCTGCAGTCCCTCTCCATGCACCTCACGTCTTTCCTCAACAGCCACGACCCGCCCCTCTCCAACGTCGTC TCCATCGAATTGGTCAACGAACCTGCACCGCCTAACGATGGTGTGCTGAAGCGGTGGTACGACGATGCGATAAGG GTTGTGCAGAGGGGTGCGCGTGGCGTGCCGGTATATATTGGGGAGTGTTGGAGGCCGGAGGTGTATGCTGAGTAT GTGGCGAGGGAGCTGAAGCAGGAGGATCCAGAAGGCCTAGTAGTACTAGACCACCACCTCTACCGTTGCTTCACC CAAGAAGACATCAACACATCCGTCTACGACCACACCGGGCGGCTGCGCGACCACAACGGCATACTCTCGACCTTT ACGCGCGTCGCTGAGCTCGTGGGTCGAGCCGGCGGGGCGCTATCCGTGTGCGAGTGGTCTGGCGGTCTCAATGAG GGGAGTTTGAAGCATGTGCGTGGGGAGGACCAACGGAGGAATGCGAGACGTGATTTTTTGAGAGCGCAGTTGGAG TTGTATGAGCGGACGTGTGCGGGGTGGTATTTTTGGACGTATAAGAAGGAGTATAGGGGTGATGTGGGGTGGTCG TTTAGGGAGGCGGTGGAGGAGGGGGTGTTTCCTATGAAGAGTGCGTAGTCACGTTTCTGCTTTGTGGTGTTAGAA CTGATGGACCTGGGTCGTGCAGATGGACGAATTGACGCTGGTATTTTGGTGAGGAAGAAGAGGTGGACGGATGCG GATGTGGAAAGGGAATGTGTGCGTAGGGTGGGGAAGATGGAGGAGGAGAGGAAGAAGGCGTATGGTAAGTCGTCT TGCGTGTTAGTTGTCATTCGCGTTTGTCCTTTCATATTTGTCCATCCACGTTTGTCCCTTCACTTCTGTCCTTCA CTTTGACCATTCACGCTTGCCCTTCACGCCATCCATCCATACCATCCAATACCCCCAACTAACTAATTCGTCCCT CGTCCCTTCACCCCACAGATGCACATAGATCCTACTGGTCCCGCATCCCGGGGCGCTACGAGCACCGCAGGTTCT GGTATGGTTTTACAGCTGGGTGGAATATGGCGTGGAGGTTTTATGATTCTATCGAACCGGATGGTGAGTAACGAT GAACGTTTCGCTTTTGTCTTGGTCGGTTTTTTTGCGTATATGTACACTATGTGGCTCCTCTCGCGCTCGAGCTGC TCGCCAATTGGGTCGTTCCTTCGTCCCTCATCGTTCTTTCCTTACTCTCATCGTTCATCCCCGTCGTTCTCTCCC TCGTTATTCTTTCTCCCATCCTCCCCCTTCCCTATCCTCCTCCTGACGATCCATCATTCTGACCGTTGTTTTACC CAACCCCTCCTCCCTTTCTTTCTTTCTTTCTCCCCCTCGGACTCGTATAGGCATAGGCAACCCAGTAAGCGAACC AGGCCCGCTACACGCCCTCGCGCGGAGAAGTTCGGTGTACCCCGTTGAGGGCCCGCATTATTGGGAGTATGAGCA TGGGTTTTTGCAGGGTGTGGGGAGTGCTAGAGGGGATTTTGAGGGGGAGTTTTGCTAAGGGGTATTCAGGAGAGA GTTTTTGTTGGTAGTGGGCGGGGTGGTGGGGAGGGAGGTTTGTGAGGTTTGTAATTGGGAAGGGGTTTGTTAGCG GGCGGGGTTGGGGAGGGAGGTTTGGTAGCGGGCTGGTTGTTGGGGAGGGGGCGTAGGTTATCTAGGCGTTGTGGG AGGGTACGTTATCTAGGAGTTGTGGGAGGGGAGGAAAGCCAGTAGAAAGAGTGAAATTTGCTTTATTTTTTTGTA GCGTAGCGTAAGCAACAAGTGGAAAGTTGGAGAGTCGCAGTTGCGTCTGGTGGTACTAGTAGGCTACTCTGGGTA TATCGTATTATGTATACTAAAGAGCGTGGACGAGGAAAGGGATGATTGTGGACTGGTAGTAGTAAGGTAGATGGA AAAAATGGATCTCCGTTCCTGCAGATGCAGAAGGGTTTG |
| Length | 3114 |