CopciAB_446910
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_446910 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 446910 |
| Uniprot id | Functional description | P21-Rho-binding domain | |
| Location | scaffold_10:1698656..1702209 | Strand | - |
| Gene length (nt) | 3554 | Transcript length (nt) | 3112 |
| CDS length (nt) | 2433 | Protein length (aa) | 810 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Saccharomyces cerevisiae | YNL298W_CLA4 |
| Aspergillus nidulans | AN8836_cla4 |
| Neurospora crassa | vel |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7264025 | 74.7 | 0 | 1203 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB18207 | 71.6 | 0 | 1199 |
| Pleurotus eryngii ATCC 90797 | Pleery1_142286 | 71.1 | 0 | 1134 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_102264 | 66.3 | 0 | 1091 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_153517 | 66.2 | 0 | 1087 |
| Flammulina velutipes | Flave_chr10AA01255 | 66 | 0 | 1060 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_130362 | 64.3 | 0 | 1049 |
| Schizophyllum commune H4-8 | Schco3_2638161 | 69.4 | 0 | 1024 |
| Pleurotus ostreatus PC15 | PleosPC15_2_34426 | 64 | 1.195E-304 | 987 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_17003 | 57.2 | 1.032E-304 | 970 |
| Auricularia subglabra | Aurde3_1_1274621 | 61.7 | 3.729E-304 | 969 |
| Pleurotus ostreatus PC9 | PleosPC9_1_87656 | 65.1 | 2.695E-304 | 957 |
| Lentinula edodes NBRC 111202 | Lenedo1_1060871 | 58.7 | 4.717E-285 | 887 |
| Lentinula edodes B17 | Lened_B_1_1_12395 | 53.3 | 2.332E-251 | 789 |
| Grifola frondosa | Grifr_OBZ74620 | 60.8 | 2.42E-221 | 702 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_446910.T0 |
| Description | P21-Rho-binding domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd13279 | PH_Cla4_Ste20 | - | 81 | 169 |
| CDD | cd01093 | CRIB_PAK_like | IPR033923 | 179 | 220 |
| CDD | cd06614 | STKc_PAK | - | 536 | 790 |
| Pfam | PF00169 | PH domain | IPR001849 | 82 | 165 |
| Pfam | PF00786 | P21-Rho-binding domain | IPR000095 | 179 | 234 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 537 | 789 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR000095 | CRIB domain |
| IPR011993 | PH-like domain superfamily |
| IPR001849 | Pleckstrin homology domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR017441 | Protein kinase, ATP binding site |
| IPR036936 | CRIB domain superfamily |
| IPR033923 | p21 activated kinase binding domain |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004674 | protein serine/threonine kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K19833 |
EggNOG
| COG category | Description |
|---|---|
| T | P21-Rho-binding domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
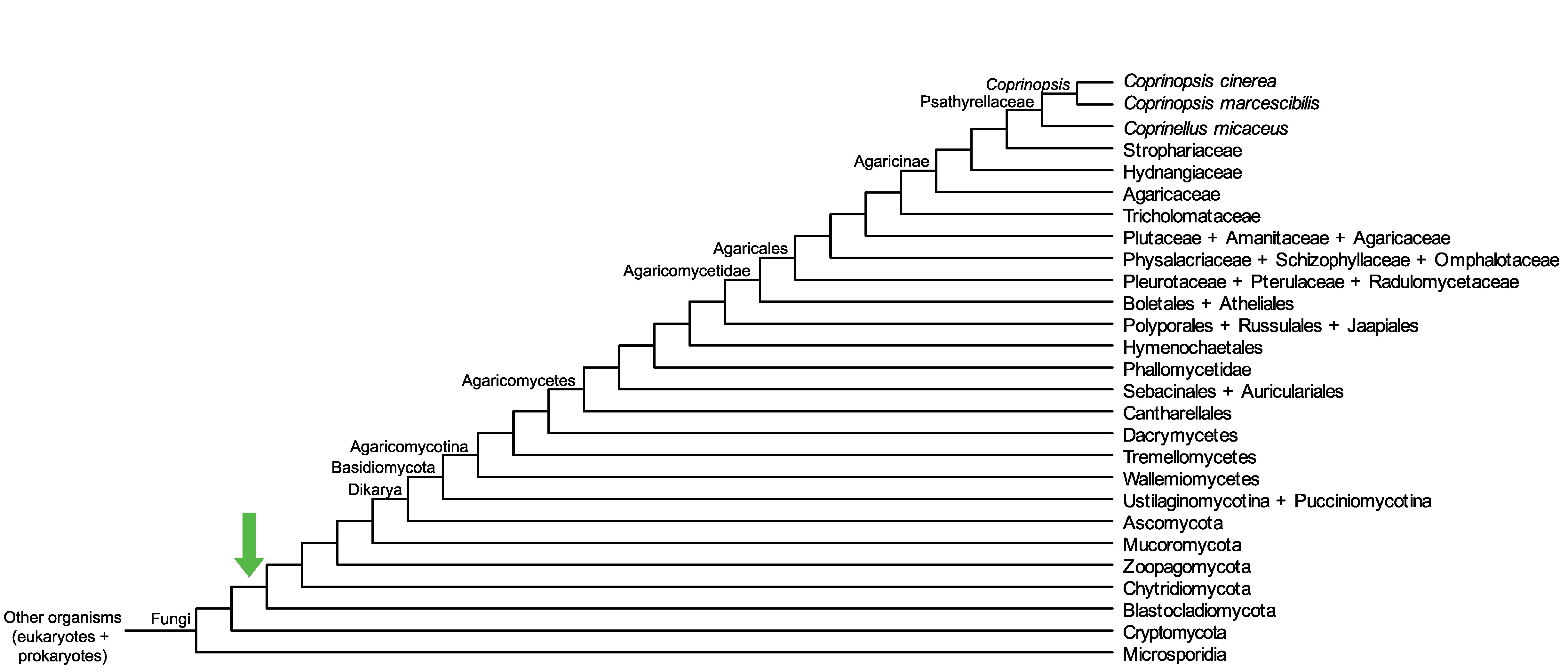
Conservation of CopciAB_446910 across fungi.
Arrow shows the origin of gene family containing CopciAB_446910.
Protein
| Sequence id | CopciAB_446910.T0 |
|---|---|
| Sequence |
>CopciAB_446910.T0 MSYHQVSANSLQPTRPAPQAPQRRNDAQPLYSSNNLLSSSGYNPSFAYSSSTPINQNYSQSYSGIGGTPNRGADD TMNQQIVRNGSVSMKEDGFGNWLWQRKWLILREQALTIHKNENSPHQTMILLRDISNIERTDLKPYCLLLETKDK RYFLSLKNDEELYGWQDDVYSRSPLMGFSNPTNFVHKVHVGFDPVSGAFTGLPEQWSKLLTKSAITREDYAKDPQ AVLDVLEFYTDRQKQELEDLSIGAPPRFNAGTGLGGLGKQPGPAEIRPLPKRNETSPPGLTGTDSDNLSSAAARA AEHVNGSHVANAISSVQGPRGPLPVPTLPQATRAAPPRPIAASRPPPQPPKDVTPSTDDLRNRVKPQGPVPLPVR NDSLPTAPGIHQSASQRSQQEQRERQQQQEQRRLEQREREREQREREREREREREEREAYRAQQERPPLAPSKSA PGTAQPATQPATGPAGALEGPPPVLPLQPKKVQIQAADKPAKPAGGVAAAAAALEKPKEKRISTMTEVQIMEKLR QVVSDEDPKLIYSKIKKVGQGASGHVYVAKELATGKKVAIKEMDLSHQPRKELIVNEILVMKESQHPNIVNFLNS YLVKNNELWVVMEYMEGGALTDVIENNTLEEDQISSICLETCKGLQHLHSQSIIHRDIKSDNVLLDAQGRVKITD FGFCAKLTDQKSKRATMVGTPYWMAPEVVKQKEYGAKVDIWSLGIMAIEMIENEPPYLDEEPLKALYLIATNGTP TLKKPEALSKELKSFLAVCLCVDVASRATAIELLDHEFLKKACQLSGLAPLLRFKTKQAS |
| Length | 810 |
Coding
| Sequence id | CopciAB_446910.T0 |
|---|---|
| Sequence |
>CopciAB_446910.T0 ATGTCTTATCACCAGGTCTCAGCCAACTCGCTCCAGCCAACAAGACCAGCACCCCAGGCACCGCAAAGACGAAAC GACGCACAACCGCTGTACTCGTCGAACAACCTGCTCTCCAGCTCTGGATACAACCCTTCTTTCGCCTACTCGTCA TCAACGCCCATAAACCAGAACTATTCACAGTCGTACTCTGGGATCGGTGGAACACCAAATCGTGGCGCAGACGAT ACCATGAACCAGCAGATTGTGCGGAACGGGAGTGTGAGTATGAAGGAGGATGGATTTGGCAACTGGCTATGGCAG AGAAAGTGGTTGATCCTGAGGGAGCAGGCCCTGACAATACACAAGAACGAAAATTCACCGCACCAAACCATGATT CTCCTTCGCGACATCTCCAACATTGAGCGCACTGATTTGAAGCCATATTGCCTGCTACTCGAGACCAAAGACAAG CGATATTTCCTTTCTTTGAAGAACGATGAGGAGCTTTATGGTTGGCAAGACGATGTGTATTCGCGAAGCCCGCTG ATGGGCTTCAGCAACCCGACGAATTTCGTCCACAAGGTCCACGTCGGTTTCGATCCTGTTTCAGGTGCCTTCACG GGTCTGCCCGAACAATGGAGCAAGCTGCTGACCAAATCTGCAATCACTCGCGAGGATTATGCCAAGGATCCCCAA GCGGTGTTGGATGTCCTAGAGTTCTATACGGATCGCCAAAAGCAGGAACTGGAAGATCTCTCAATAGGTGCTCCT CCCCGCTTCAACGCTGGGACCGGCCTGGGAGGCCTTGGGAAGCAACCGGGCCCCGCTGAAATCCGCCCACTGCCC AAGCGCAACGAGACCTCACCGCCTGGCCTTACTGGCACGGATAGCGATAACCTTTCCAGCGCGGCCGCTCGTGCA GCTGAACACGTCAACGGATCTCATGTCGCGAACGCCATCTCCTCTGTGCAAGGTCCCCGAGGCCCTCTGCCTGTC CCCACCCTTCCTCAAGCCACCCGCGCAGCACCACCCCGTCCCATCGCGGCCTCTCGGCCACCTCCCCAGCCTCCC AAGGATGTCACTCCATCCACCGACGATCTCCGCAATCGCGTGAAGCCGCAAGGCCCCGTCCCTCTCCCCGTCCGC AACGATTCCCTCCCTACTGCCCCTGGAATCCATCAATCCGCCAGCCAGAGAAGCCAGCAAGAGCAACGCGAGCGC CAGCAGCAGCAGGAGCAGCGTCGGCTGGAGCAACGCGAGAGAGAGCGAGAGCAGCGTGAACGTGAGCGCGAGCGC GAGCGAGAGAGAGAGGAAAGAGAGGCGTATAGGGCTCAGCAAGAGCGACCCCCTCTCGCGCCTTCCAAGTCTGCT CCTGGGACTGCTCAGCCGGCGACGCAGCCTGCTACAGGCCCCGCTGGTGCCCTTGAGGGACCACCGCCGGTTCTT CCCCTCCAGCCCAAGAAAGTGCAGATCCAAGCGGCTGATAAGCCAGCGAAACCTGCCGGCGGTGTTGCTGCTGCC GCTGCGGCATTGGAGAAGCCCAAGGAGAAGAGGATCAGTACCATGACTGAGGTTCAGATCATGGAGAAACTGCGT CAAGTGGTATCAGACGAGGACCCCAAGCTTATCTACAGCAAGATTAAGAAAGTCGGACAAGGCGCGTCTGGCCAC GTCTATGTTGCGAAGGAGCTTGCCACCGGAAAGAAGGTTGCGATCAAGGAGATGGATCTCTCACACCAGCCGCGT AAAGAGCTCATCGTCAATGAGATCTTGGTGATGAAGGAGTCGCAGCATCCTAACATTGTCAACTTCTTGAACTCG TATCTCGTGAAGAACAATGAGCTATGGGTGGTCATGGAGTACATGGAGGGAGGTGCGTTGACGGATGTTATTGAG AATAACACGTTGGAGGAAGATCAGATCTCGAGTATCTGCTTGGAGACTTGCAAGGGTTTGCAACATCTGCATAGC CAGAGCATCATTCACCGTGACATCAAGTCCGACAACGTCCTTCTCGATGCTCAGGGTCGGGTCAAGATCACTGAC TTTGGCTTCTGCGCCAAACTAACGGACCAGAAGTCGAAGCGGGCGACCATGGTTGGTACTCCCTACTGGATGGCT CCCGAAGTCGTCAAGCAGAAGGAGTACGGTGCCAAGGTCGATATTTGGTCGCTTGGTATCATGGCGATTGAGATG ATTGAGAACGAGCCTCCCTATCTCGACGAAGAACCTCTCAAGGCGCTGTATCTGATCGCTACCAACGGAACTCCC ACTCTGAAGAAGCCTGAGGCTCTGAGCAAGGAGCTCAAGAGCTTCTTGGCGGTGTGTCTCTGCGTGGATGTTGCT AGTCGGGCGACTGCCATTGAACTTTTGGATCACGAGTTCTTGAAAAAGGCGTGCCAATTGTCCGGGCTAGCTCCA CTTTTGCGCTTCAAGACCAAGCAAGCGTCCTAA |
| Length | 2433 |
Transcript
| Sequence id | CopciAB_446910.T0 |
|---|---|
| Sequence |
>CopciAB_446910.T0 AGCTCATCGCGCTCGACGACGACCACGACGACCAATCTCACGATCGTAACGACGCTCTCAACGCTCCAAGTGACG CTCACAAGCGTCCAACGTCGTCTCTGAGCCACCTCAGTCCCAGCTCGCCATCGCGTTCGATCCTGGACCAGTTGC TTCCTTGGTGACCGTCGACGGCGTCGGCCAAACTCCCACTCGATATCGTCCAGCCTCGTCCGACGGCCATGTCTT ATCACCAGGTCTCAGCCAACTCGCTCCAGCCAACAAGACCAGCACCCCAGGCACCGCAAAGACGAAACGACGCAC AACCGCTGTACTCGTCGAACAACCTGCTCTCCAGCTCTGGATACAACCCTTCTTTCGCCTACTCGTCATCAACGC CCATAAACCAGAACTATTCACAGTCGTACTCTGGGATCGGTGGAACACCAAATCGTGGCGCAGACGATACCATGA ACCAGCAGATTGTGCGGAACGGGAGTGTGAGTATGAAGGAGGATGGATTTGGCAACTGGCTATGGCAGAGAAAGT GGTTGATCCTGAGGGAGCAGGCCCTGACAATACACAAGAACGAAAATTCACCGCACCAAACCATGATTCTCCTTC GCGACATCTCCAACATTGAGCGCACTGATTTGAAGCCATATTGCCTGCTACTCGAGACCAAAGACAAGCGATATT TCCTTTCTTTGAAGAACGATGAGGAGCTTTATGGTTGGCAAGACGATGTGTATTCGCGAAGCCCGCTGATGGGCT TCAGCAACCCGACGAATTTCGTCCACAAGGTCCACGTCGGTTTCGATCCTGTTTCAGGTGCCTTCACGGGTCTGC CCGAACAATGGAGCAAGCTGCTGACCAAATCTGCAATCACTCGCGAGGATTATGCCAAGGATCCCCAAGCGGTGT TGGATGTCCTAGAGTTCTATACGGATCGCCAAAAGCAGGAACTGGAAGATCTCTCAATAGGTGCTCCTCCCCGCT TCAACGCTGGGACCGGCCTGGGAGGCCTTGGGAAGCAACCGGGCCCCGCTGAAATCCGCCCACTGCCCAAGCGCA ACGAGACCTCACCGCCTGGCCTTACTGGCACGGATAGCGATAACCTTTCCAGCGCGGCCGCTCGTGCAGCTGAAC ACGTCAACGGATCTCATGTCGCGAACGCCATCTCCTCTGTGCAAGGTCCCCGAGGCCCTCTGCCTGTCCCCACCC TTCCTCAAGCCACCCGCGCAGCACCACCCCGTCCCATCGCGGCCTCTCGGCCACCTCCCCAGCCTCCCAAGGATG TCACTCCATCCACCGACGATCTCCGCAATCGCGTGAAGCCGCAAGGCCCCGTCCCTCTCCCCGTCCGCAACGATT CCCTCCCTACTGCCCCTGGAATCCATCAATCCGCCAGCCAGAGAAGCCAGCAAGAGCAACGCGAGCGCCAGCAGC AGCAGGAGCAGCGTCGGCTGGAGCAACGCGAGAGAGAGCGAGAGCAGCGTGAACGTGAGCGCGAGCGCGAGCGAG AGAGAGAGGAAAGAGAGGCGTATAGGGCTCAGCAAGAGCGACCCCCTCTCGCGCCTTCCAAGTCTGCTCCTGGGA CTGCTCAGCCGGCGACGCAGCCTGCTACAGGCCCCGCTGGTGCCCTTGAGGGACCACCGCCGGTTCTTCCCCTCC AGCCCAAGAAAGTGCAGATCCAAGCGGCTGATAAGCCAGCGAAACCTGCCGGCGGTGTTGCTGCTGCCGCTGCGG CATTGGAGAAGCCCAAGGAGAAGAGGATCAGTACCATGACTGAGGTTCAGATCATGGAGAAACTGCGTCAAGTGG TATCAGACGAGGACCCCAAGCTTATCTACAGCAAGATTAAGAAAGTCGGACAAGGCGCGTCTGGCCACGTCTATG TTGCGAAGGAGCTTGCCACCGGAAAGAAGGTTGCGATCAAGGAGATGGATCTCTCACACCAGCCGCGTAAAGAGC TCATCGTCAATGAGATCTTGGTGATGAAGGAGTCGCAGCATCCTAACATTGTCAACTTCTTGAACTCGTATCTCG TGAAGAACAATGAGCTATGGGTGGTCATGGAGTACATGGAGGGAGGTGCGTTGACGGATGTTATTGAGAATAACA CGTTGGAGGAAGATCAGATCTCGAGTATCTGCTTGGAGACTTGCAAGGGTTTGCAACATCTGCATAGCCAGAGCA TCATTCACCGTGACATCAAGTCCGACAACGTCCTTCTCGATGCTCAGGGTCGGGTCAAGATCACTGACTTTGGCT TCTGCGCCAAACTAACGGACCAGAAGTCGAAGCGGGCGACCATGGTTGGTACTCCCTACTGGATGGCTCCCGAAG TCGTCAAGCAGAAGGAGTACGGTGCCAAGGTCGATATTTGGTCGCTTGGTATCATGGCGATTGAGATGATTGAGA ACGAGCCTCCCTATCTCGACGAAGAACCTCTCAAGGCGCTGTATCTGATCGCTACCAACGGAACTCCCACTCTGA AGAAGCCTGAGGCTCTGAGCAAGGAGCTCAAGAGCTTCTTGGCGGTGTGTCTCTGCGTGGATGTTGCTAGTCGGG CGACTGCCATTGAACTTTTGGATCACGAGTTCTTGAAAAAGGCGTGCCAATTGTCCGGGCTAGCTCCACTTTTGC GCTTCAAGACCAAGCAAGCGTCCTAAACGCTTGTGTTGGTCGTTTATGATTGATTGATTCGTTCTTTCTTCGTCT ATCTGTGTTTCTTCTTATAGTTGTTTGTTTGTTTTTGTTCCACCCATATACCGATATTCAAGATTTAAGGATACG ACCTCCCTCACCATGAACTATCCCCCCTAATGCTTCGTCGTCTTTCTGTCTTGTTTCTTTAAACCTCATGTGTGC CCTTCATTCTACCGTTGCCTGGAATTTACATGTTTCACTCGGTGCCCCCCTTTTTCGACGTTGTCTCCTTTTAAG CAGGTGCTTTCTGTAAATTCTTCTTCTCTATCTTTCCCTTCCTCCTTCCTTTTGCACATTTTGTTCTTTTACAAT TACGAGATCCCTTTTTTTTGTCTTTTATTATTACCCCCCTCTTGTCTGTTTGTTTGCTACACCCTCCATGTATTT GTTGCCGAATGTGTAACCTCTTTTTTTCGCCTGTTTC |
| Length | 3112 |
Gene
| Sequence id | CopciAB_446910.T0 |
|---|---|
| Sequence |
>CopciAB_446910.T0 AGCTCATCGCGCTCGACGACGACCACGACGACCAATCTCACGATCGTAACGACGCTCTCAACGCTCCAAGTGACG CTCACAAGCGTCCAACGTCGTCTCTGAGCCACCTCAGTCCCAGCTCGCCATCGCGTTCGATCCTGGACCAGTTGC TTCCTTGGTGACCGTCGACGGCGTCGGCCAAACTCCCACTCGATATCGTCCAGCCTCGTCCGACGGCCATGTCTT ATCACCAGGTCTCAGCCAACTCGCTCCAGCCAACAAGACCAGCACCCCAGGCACCGCAAAGACGAAACGACGCAC AACCGCTGTACTCGTCGAACAACCTGCTCTCCAGCTCTGGATACAACCCTTCTTTCGCCTACTCGTCATCAACGC CCATAAACCAGAACTATTCACAGTCGTACTCTGGGATCGGTGGAACACCAAATCGTGGCGCAGACGATACCATGA ACCAGCAGATTGTGCGGAACGGGAGTGTGAGTATGAAGGAGGATGGATTTGGCAACTGGCTATGGCAGAGAAAGT GGTTGATCCTGAGGGAGCAGGCCCTGACAATACACAAGAACGAAGTGAGTAACGAATATCCTCTTTTCAAAGGAT ATGCTAATCAGTGCTGTTAACAGAATTCACCGCACCAAACCATGATTCTCCTTCGCGACATCTCCAACATTGAGC GCACTGATTTGAAGCCATATTGCCTGCTACTCGAGACCAAAGACAAGCGATATTTCCTTTCTTTGAAGAACGATG AGGAGCTTTATGGTTGGCAAGACGATGTGTATTCGCGAAGCCCGCTGATGGGCTTCAGCAACCCGACGAATTTCG TCCACAAGGTCCACGTCGGTTTCGATCCTGTTTCAGGTGCCTTCACGGTAAGCCTTCATCAGAACGTTCTGCCGT GCCCCTGGTCCTGACTCAATTAATTCAAGGGTCTGCCCGAACAATGGAGCAAGCTGCTGACCAAATCTGCAATCA CTCGCGAGGATTATGCCAAGGATCCCCAAGCGGTGTTGGATGTCCTAGAGTTCTATACGGATCGCCAAAAGCAGG AACTGGAAGATCTCTCAATAGGTGCGTGGTCACATACCCGTGCTCGGGATCCCCTCTGATCCCACGCTATCATTA CTAGGTGCTCCTCCCCGCTTCAACGCTGGGACCGGCCTGGGAGGCCTTGGGAAGCAACCGGGCCCCGCTGAAATC CGCCCACTGCCCAAGCGCAACGAGACCTCACCGCCTGGCCTTACTGGCACGGATAGCGATAACCTTTCCAGCGCG GCCGCTCGTGCAGCTGAACACGTCAACGGATCTCATGTCGCGAACGCCATCTCCTCTGTGCAAGGTCCCCGAGGC CCTCTGCCTGTCCCCACCCTTCCTCAAGCCACCCGCGCAGCACCACCCCGTCCCATCGCGGCCTCTCGGCCACCT CCCCAGCCTCCCAAGGATGTCACTCCATCCACCGACGATCTCCGCAATCGCGTGAAGCCGCAAGGCCCCGTCCCT CTCCCCGTCCGCAACGATTCCCTCCCTACTGCCCCTGGAATCCATCAATCCGCCAGCCAGAGAAGCCAGCAAGAG CAACGCGAGCGCCAGCAGCAGCAGGAGCAGCGTCGGCTGGAGCAACGCGAGAGAGAGCGAGAGCAGCGTGAACGT GAGCGCGAGCGCGAGCGAGAGAGAGAGGAAAGAGAGGCGTATAGGGCTCAGCAAGAGCGACCCCCTCTCGCGCCT TCCAAGTCTGCTCCTGGGACTGCTCAGCCGGCGACGCAGCCTGCTACAGGCCCCGCTGGTGCCCTTGAGGGACCA CCGCCGGTTCTTCCCCTCCAGCCCAAGAAAGTGCAGATCCAAGCGGCTGATAAGCCAGCGAAACCTGCCGGCGGT GTTGCTGCTGCCGCTGCGGCATTGGAGAAGCCCAAGGAGAAGAGGATCAGTACCATGACTGAGGTTCAGATCATG GAGAAACTGCGTCAAGTGGTATCAGACGAGGACCCCAAGCTTATCTACAGCAAGATTAAGAAAGTCGGACAAGGG TGAGTAAACACGAGGCCAATCGTTTCTTTTTGTTATTCCCTCGTTGGTACTTTTTGTTGGAGCGATGTCACTGAC GAGACGTTTTTTGTGACTTCTAGCGCGTCTGGCCACGTCTATGTTGCGAAGGAGCTTGCCACCGGAAAGAAGGTT GCGATCAAGGAGATGGATCTCTCACACCAGCCGCGTAAAGAGCTCATCGTCAATGAGATCTTGGTGATGAAGGAG TCGCAGCATCCTAACATTGTCAACTTCTTGAACTCGTATCTCGTGAAGAACAATGAGCTATGGGTGGTCATGGAG TACATGGAGGGAGGTGCGTTGACGGATGTTATTGAGAATAACACGTTGGAGGAAGATCAGATCTCGAGTATCTGC TTGGAGGTGCGTGGTTGGCGCTATGAGATAGGAACGTAACTGACGTTGACGTGACTGTAGACTTGCAAGGGTTTG CAACATCTGCATAGCCAGAGCATCATTCACCGTGACATCAAGTCCGACAACGTCCTTCTCGATGCTCAGGGTCGG GTCAAGATCAGTGCGTGTGCCCCCTGTGACATTTATCGCCACATTCTGACGTTTTCTTCAACAGCTGACTTTGGC TTCTGCGCCAAACTAACGGACCAGAAGTCGAAGCGGGCGACCATGGTTGGTACTCCCTACTGGATGGCTCCCGAA GTCGTCAAGCAGAAGGAGTACGGTGCCAAGGTCGATATTTGGTCGCTTGGTATCATGGCGATTGAGATGATTGAG AACGAGCCTCCCTATCTCGACGAAGAACCTCTCAAGGCGCTGTATCTGATCGCTACCAACGGAACTCCCACTCTG AAGAAGCCTGAGGCTCTGAGCAAGGAGCTCAAGAGCTTCTTGGCGGTGTGTCTCTGCGTGGATGTTGCTAGTCGG GCGACTGCCATTGAACTTTTGGATGTGAGTGCTCTTTTTTTCCTTTGAGGTGGGCCGATGCTGATCGCCGCGGTA CCTGCCTTTTTACAGCACGAGTTCTTGAAAAAGGCGTGCCAATTGTCCGGGCTAGCTCCACTTTTGCGCTTCAAG ACCAAGCAAGCGTCCTAAACGCTTGTGTTGGTCGTTTATGATTGATTGATTCGTTCTTTCTTCGTCTATCTGTGT TTCTTCTTATAGTTGTTTGTTTGTTTTTGTTCCACCCATATACCGATATTCAAGATTTAAGGATACGACCTCCCT CACCATGAACTATCCCCCCTAATGCTTCGTCGTCTTTCTGTCTTGTTTCTTTAAACCTCATGTGTGCCCTTCATT CTACCGTTGCCTGGAATTTACATGTTTCACTCGGTGCCCCCCTTTTTCGACGTTGTCTCCTTTTAAGCAGGTGCT TTCTGTAAATTCTTCTTCTCTATCTTTCCCTTCCTCCTTCCTTTTGCACATTTTGTTCTTTTACAATTACGAGAT CCCTTTTTTTTGTCTTTTATTATTACCCCCCTCTTGTCTGTTTGTTTGCTACACCCTCCATGTATTTGTTGCCGA ATGTGTAACCTCTTTTTTTCGCCTGTTTC |
| Length | 3554 |