CopciAB_447354
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_447354 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 447354 |
| Uniprot id | Functional description | other FunK1 protein kinase | |
| Location | scaffold_7:2012870..2015699 | Strand | - |
| Gene length (nt) | 2830 | Transcript length (nt) | 2575 |
| CDS length (nt) | 2406 | Protein length (aa) | 801 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_132224 | 30.1 | 3.198E-66 | 243 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_122759 | 27 | 2.262E-54 | 206 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1249338 | 23.9 | 1.027E-21 | 101 |
| Auricularia subglabra | Aurde3_1_1411434 | 23.5 | 1.849E-18 | 91 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_447354.T0 |
| Description | other FunK1 protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 373 | 585 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR040976 | Fungal-type protein kinase |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| G | other FunK1 protein kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
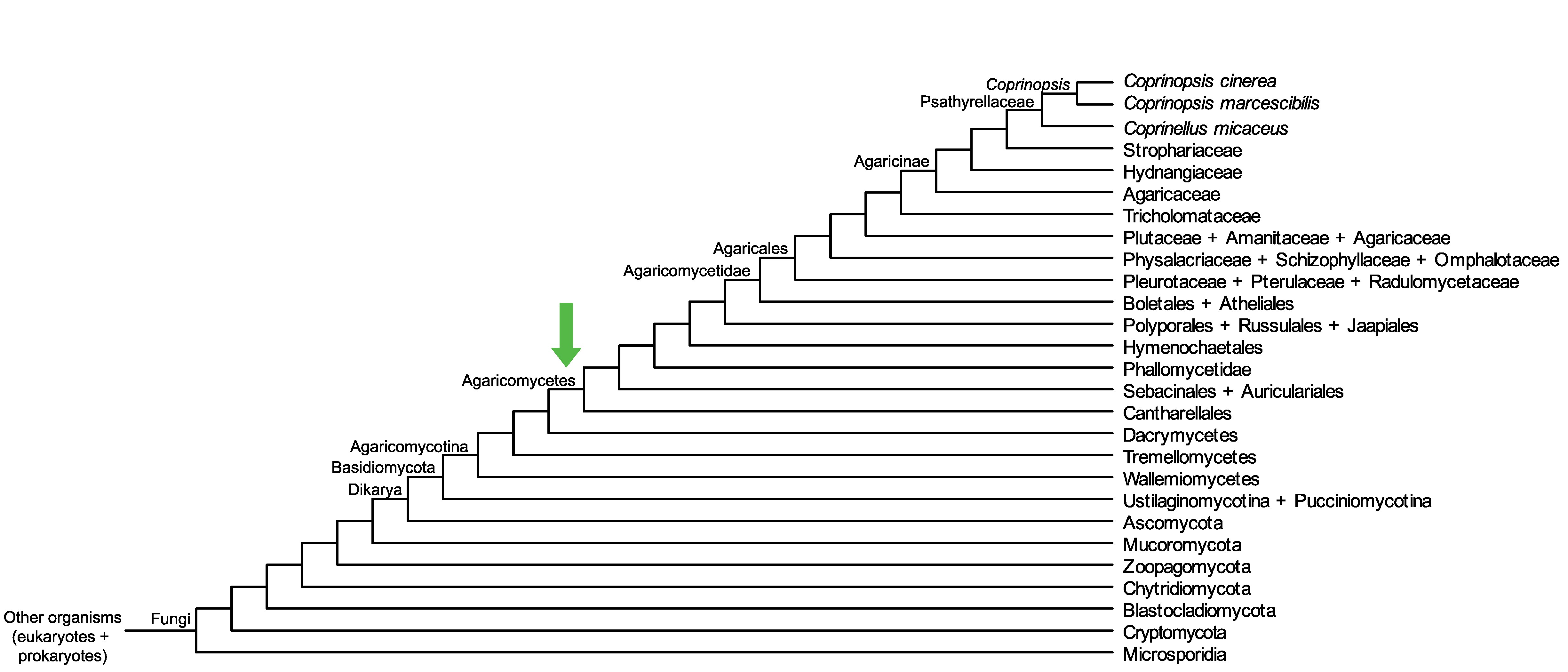
Conservation of CopciAB_447354 across fungi.
Arrow shows the origin of gene family containing CopciAB_447354.
Protein
| Sequence id | CopciAB_447354.T0 |
|---|---|
| Sequence |
>CopciAB_447354.T0 MSASTPVIVRQAPFALNNRPERQIFAAYTPLSVHIDDASPPFLAKVNLTIGDPALKLDGTPQFNSLNEFFKDVNA VCEQMRAAFNFKRLLIFKETQNTIQQGHVSDTVAKPDLVAGFEDHWYDGRAPATQKKSEAGKGGSADPHKEDLKS HMLWPLIRLAGEIALSSETQGEQIQHAGTYLDLLLEARPDFKAAFGLLLSENSVFVLVGETGAGIRLFSFSWGSR DLRTAMRAMVYRLYNPGEWANTSIEACFRTASTKDKPPSCVYNLTFPCSDSQKAPVYKNYFCRYGKSSFGTRTHV FVHGGPQPQTINGFPLRAIKIQLCRDSGRFNEIMVLNHIHKNAVLPGVIRLLCSHSSSHLFGHRKEIWLGLDKEG EPFMAIKTVREMLMVAYDVLEITRMLFARRRVLHRDISKGNVMYISRRTMTQIIKTIKAPDATAVGKNQGDGSCY CFVRYLLGESIDSMETSALLIDFNYSEIVPEDESTGGHAVGARTGTPMFMARAVQNGEPIKLEPYHTVTLVPIPS SAHLGTYEKNFPDRISRFRELDRKENIEANEVGEPTHEILLPWRQRLYHDAESVYWLMYYWGLLAKPSTSIKEKG AEDFDPGIPANIWTELHAGRKTLSAHANDTFHPSYSPLLDLFVQMRAYLLTDPHWFPHDSPRAEPDYIHECFQRL ILRFLKDNVDKPFMELKKSDHERLPFAVPGIPARSSSTTDTYLSDQRAQRPETPPIPREPFAVNYVAPPVVAGFQ STDNTPIQRTMSLPGKEGDEDSVIEQLVFPQGSSRQGPGRSQESNNDDVFT |
| Length | 801 |
Coding
| Sequence id | CopciAB_447354.T0 |
|---|---|
| Sequence |
>CopciAB_447354.T0 ATGTCCGCGTCGACCCCTGTTATCGTGCGGCAAGCGCCCTTCGCGCTGAACAACAGACCTGAACGCCAAATCTTC GCTGCGTACACACCGCTATCTGTGCACATCGACGATGCGAGTCCACCGTTCTTGGCAAAAGTCAACCTCACGATT GGGGATCCTGCTCTGAAGCTTGATGGAACCCCACAATTTAACTCGCTGAACGAGTTCTTCAAAGACGTGAATGCC GTCTGTGAGCAGATGAGGGCAGCATTCAACTTCAAGCGTCTCCTCATCTTCAAAGAGACACAAAATACCATCCAG CAAGGACATGTTTCAGATACAGTTGCAAAACCCGACTTGGTTGCCGGGTTCGAAGATCATTGGTACGATGGTCGA GCTCCTGCCACTCAGAAGAAATCCGAGGCCGGCAAGGGCGGCAGTGCGGACCCTCATAAAGAGGACCTCAAGTCC CACATGCTATGGCCTCTCATCAGGCTTGCCGGGGAGATTGCTTTGTCAAGCGAAACCCAGGGAGAACAAATCCAA CATGCTGGCACCTACCTCGACCTTCTGCTTGAGGCTCGGCCCGATTTCAAAGCCGCCTTCGGTCTTCTGCTCAGC GAAAACAGCGTGTTCGTGCTCGTTGGGGAGACGGGTGCGGGTATCCGCCTTTTCTCGTTTTCCTGGGGAAGCCGC GACCTGCGGACGGCGATGCGCGCTATGGTATACCGACTATACAACCCAGGGGAGTGGGCGAACACCTCTATCGAG GCATGCTTCAGGACAGCGTCGACCAAAGACAAGCCTCCTTCGTGTGTATACAACCTCACTTTCCCGTGCTCGGAT TCCCAAAAGGCGCCCGTATACAAGAATTACTTCTGTCGATATGGAAAAAGTTCCTTCGGTACTCGGACGCACGTT TTCGTCCACGGTGGCCCCCAGCCTCAAACTATCAACGGTTTTCCTCTCAGGGCCATCAAGATACAGCTCTGTCGC GACTCGGGTCGTTTTAACGAAATCATGGTGCTGAACCACATACACAAAAATGCCGTCCTCCCTGGGGTGATCCGT TTGCTCTGTTCCCATAGTTCCTCGCACCTCTTCGGACATAGGAAAGAAATCTGGTTGGGATTAGACAAAGAGGGC GAACCTTTCATGGCGATCAAGACCGTTCGGGAGATGCTCATGGTGGCCTATGATGTCCTAGAAATCACCCGCATG CTCTTCGCCCGACGCAGGGTCCTGCACCGCGACATAAGCAAAGGGAATGTCATGTATATTTCCCGGCGCACAATG ACACAGATCATTAAGACCATTAAGGCCCCCGACGCGACTGCCGTAGGAAAGAATCAAGGAGACGGTTCCTGCTAC TGTTTCGTCAGGTATTTGCTCGGCGAAAGCATCGATTCTATGGAGACTTCCGCCCTGTTAATCGATTTCAACTAT AGTGAAATAGTGCCTGAGGACGAGTCAACAGGGGGACATGCGGTAGGTGCCCGAACAGGAACTCCCATGTTCATG GCCAGAGCCGTTCAAAATGGAGAGCCGATCAAGCTCGAGCCTTATCACACGGTCACCTTGGTCCCGATACCAAGC TCTGCACACTTGGGTACCTACGAGAAAAACTTCCCGGACCGCATTTCTCGGTTCCGTGAGCTGGACAGGAAGGAG AATATTGAGGCAAACGAAGTCGGGGAGCCCACGCACGAGATTCTCCTCCCCTGGAGACAACGACTGTATCATGAT GCCGAATCGGTCTATTGGCTCATGTACTATTGGGGCTTGCTGGCAAAGCCTTCGACCAGTATCAAGGAGAAAGGG GCGGAAGATTTCGACCCGGGCATCCCTGCGAACATCTGGACAGAATTGCACGCGGGCCGCAAGACGCTCTCTGCT CACGCGAACGACACATTCCATCCCTCCTACAGCCCTCTTCTTGATCTCTTCGTGCAAATGAGGGCCTACCTGCTG ACTGATCCTCATTGGTTCCCGCACGACTCTCCCAGGGCTGAGCCGGATTACATCCACGAATGTTTTCAGCGGCTT ATTCTCAGATTTCTAAAGGACAACGTCGATAAGCCATTCATGGAATTGAAGAAGTCGGATCATGAGCGCCTCCCC TTCGCTGTTCCCGGGATCCCTGCTCGTTCCTCCTCGACAACTGATACCTACCTGTCGGATCAGCGAGCTCAAAGG CCTGAAACTCCTCCAATCCCCCGTGAGCCGTTCGCTGTCAATTATGTGGCACCCCCAGTCGTGGCTGGCTTCCAA TCGACCGATAATACCCCGATTCAGCGCACGATGTCCCTCCCAGGCAAAGAGGGCGATGAAGACAGTGTCATTGAA CAGCTTGTTTTCCCCCAAGGTTCGAGCCGCCAGGGTCCTGGTCGATCCCAAGAGTCGAACAACGATGATGTCTTC ACTTGA |
| Length | 2406 |
Transcript
| Sequence id | CopciAB_447354.T0 |
|---|---|
| Sequence |
>CopciAB_447354.T0 AAGTTCGCGTCGCGCGACAATGTCCGCGTCGACCCCTGTTATCGTGCGGCAAGCGCCCTTCGCGCTGAACAACAG ACCTGAACGCCAAATCTTCGCTGCGTACACACCGCTATCTGTGCACATCGACGATGCGAGTCCACCGTTCTTGGC AAAAGTCAACCTCACGATTGGGGATCCTGCTCTGAAGCTTGATGGAACCCCACAATTTAACTCGCTGAACGAGTT CTTCAAAGACGTGAATGCCGTCTGTGAGCAGATGAGGGCAGCATTCAACTTCAAGCGTCTCCTCATCTTCAAAGA GACACAAAATACCATCCAGCAAGGACATGTTTCAGATACAGTTGCAAAACCCGACTTGGTTGCCGGGTTCGAAGA TCATTGGTACGATGGTCGAGCTCCTGCCACTCAGAAGAAATCCGAGGCCGGCAAGGGCGGCAGTGCGGACCCTCA TAAAGAGGACCTCAAGTCCCACATGCTATGGCCTCTCATCAGGCTTGCCGGGGAGATTGCTTTGTCAAGCGAAAC CCAGGGAGAACAAATCCAACATGCTGGCACCTACCTCGACCTTCTGCTTGAGGCTCGGCCCGATTTCAAAGCCGC CTTCGGTCTTCTGCTCAGCGAAAACAGCGTGTTCGTGCTCGTTGGGGAGACGGGTGCGGGTATCCGCCTTTTCTC GTTTTCCTGGGGAAGCCGCGACCTGCGGACGGCGATGCGCGCTATGGTATACCGACTATACAACCCAGGGGAGTG GGCGAACACCTCTATCGAGGCATGCTTCAGGACAGCGTCGACCAAAGACAAGCCTCCTTCGTGTGTATACAACCT CACTTTCCCGTGCTCGGATTCCCAAAAGGCGCCCGTATACAAGAATTACTTCTGTCGATATGGAAAAAGTTCCTT CGGTACTCGGACGCACGTTTTCGTCCACGGTGGCCCCCAGCCTCAAACTATCAACGGTTTTCCTCTCAGGGCCAT CAAGATACAGCTCTGTCGCGACTCGGGTCGTTTTAACGAAATCATGGTGCTGAACCACATACACAAAAATGCCGT CCTCCCTGGGGTGATCCGTTTGCTCTGTTCCCATAGTTCCTCGCACCTCTTCGGACATAGGAAAGAAATCTGGTT GGGATTAGACAAAGAGGGCGAACCTTTCATGGCGATCAAGACCGTTCGGGAGATGCTCATGGTGGCCTATGATGT CCTAGAAATCACCCGCATGCTCTTCGCCCGACGCAGGGTCCTGCACCGCGACATAAGCAAAGGGAATGTCATGTA TATTTCCCGGCGCACAATGACACAGATCATTAAGACCATTAAGGCCCCCGACGCGACTGCCGTAGGAAAGAATCA AGGAGACGGTTCCTGCTACTGTTTCGTCAGGTATTTGCTCGGCGAAAGCATCGATTCTATGGAGACTTCCGCCCT GTTAATCGATTTCAACTATAGTGAAATAGTGCCTGAGGACGAGTCAACAGGGGGACATGCGGTAGGTGCCCGAAC AGGAACTCCCATGTTCATGGCCAGAGCCGTTCAAAATGGAGAGCCGATCAAGCTCGAGCCTTATCACACGGTCAC CTTGGTCCCGATACCAAGCTCTGCACACTTGGGTACCTACGAGAAAAACTTCCCGGACCGCATTTCTCGGTTCCG TGAGCTGGACAGGAAGGAGAATATTGAGGCAAACGAAGTCGGGGAGCCCACGCACGAGATTCTCCTCCCCTGGAG ACAACGACTGTATCATGATGCCGAATCGGTCTATTGGCTCATGTACTATTGGGGCTTGCTGGCAAAGCCTTCGAC CAGTATCAAGGAGAAAGGGGCGGAAGATTTCGACCCGGGCATCCCTGCGAACATCTGGACAGAATTGCACGCGGG CCGCAAGACGCTCTCTGCTCACGCGAACGACACATTCCATCCCTCCTACAGCCCTCTTCTTGATCTCTTCGTGCA AATGAGGGCCTACCTGCTGACTGATCCTCATTGGTTCCCGCACGACTCTCCCAGGGCTGAGCCGGATTACATCCA CGAATGTTTTCAGCGGCTTATTCTCAGATTTCTAAAGGACAACGTCGATAAGCCATTCATGGAATTGAAGAAGTC GGATCATGAGCGCCTCCCCTTCGCTGTTCCCGGGATCCCTGCTCGTTCCTCCTCGACAACTGATACCTACCTGTC GGATCAGCGAGCTCAAAGGCCTGAAACTCCTCCAATCCCCCGTGAGCCGTTCGCTGTCAATTATGTGGCACCCCC AGTCGTGGCTGGCTTCCAATCGACCGATAATACCCCGATTCAGCGCACGATGTCCCTCCCAGGCAAAGAGGGCGA TGAAGACAGTGTCATTGAACAGCTTGTTTTCCCCCAAGGTTCGAGCCGCCAGGGTCCTGGTCGATCCCAAGAGTC GAACAACGATGATGTCTTCACTTGAGGTGCAGCTTCCCTCATTCTTTCTTCCCCTCTGAATTGCGCTTTCACTAT GCTGTCATCGCTGCCGTTGTTACTTCATGTTGATGTGTTTTCCGGTTTCGATACCGATTTCCGTGTACAATATAC GTAATGAGTATTTCAGCGTTGTCGA |
| Length | 2575 |
Gene
| Sequence id | CopciAB_447354.T0 |
|---|---|
| Sequence |
>CopciAB_447354.T0 AAGTTCGCGTCGCGCGACAATGTCCGCGTCGACCCCTGTTATCGTGCGGCAAGCGCCCTTCGCGCTGAACAACAG ACCTGAACGCCAAATCTTCGCTGCGTACACACCGCTATCTGTGCACATCGACGATGCGAGTCCACCGTTCTTGGC AAAAGTCAACCTCACGATTGGGGATCCTGCTCTGAAGCTTGATGGAACCCCACAATTTAACTCGCTGAACGAGTT CTTCAAAGACGTGAATGCCGTCTGTGAGCAGATGAGGGCAGGTGCGTACTTTTCAAGTTCGTTGCCACTGACTCG GATCTGATCTGAACACCAGCATTCAACTTCAAGCGTCTCCTCATCTTCAAAGAGACACAAAATACCATCCAGCAA GGACATGTTTCAGATACAGTTGCAAAACCCGACTTGGTTGCCGGGTTCGAAGATCATTGGTACGATGGTCGAGCT CCTGCCACTCAGAAGAAATCCGAGGCCGGCAAGGGCGGCAGTGCGGACCCTCATAAAGAGGACCTCAAGTCCCAC ATGCTATGGCCTCTCATCAGGCTTGCCGGGGAGATTGCTTTGTCAAGCGAAACCCAGGGAGAACAAATCCAACAT GCTGGCACCTACCTCGACCTTCTGCTTGAGGCTCGGCCCGATTTCAAAGCCGCCTTCGGTCTTCTGCTCAGCGAA AACAGCGTGTTCGTGCTCGTTGGGGAGACGGGTGCGGGTATCCGCCTTTTCTCGTTTTCCTGGGGAAGCCGCGAC CTGCGGACGGCGATGCGCGCTATGGTATACCGACTATACAACCCAGGGGAGTGGGCGAACACCTCTATCGAGGCA TGCTTCAGGACAGCGTCGACCAAAGACAAGCCTCCTTCGTGTGTATACAACCTCACTTTCCCGTGCTCGGATTCC CAAAAGGCGCCCGTATACAAGAATTACTTCTGTCGATATGGAAAAAGTTCCTTCGGTACTCGGACGCACGTTTTC GTCCACGGTGGCCCCCAGCCTCAAACTATCAACGGTTTTCCTCTCAGGGCCATCAAGATACAGCTCTGTCGCGAC TCGGGTCGTTTTAACGAAATCATGGTGCTGAACCACATACACAAAAATGCCGTCCTCCCTGGGGTGATCCGTTTG CTCTGTTCCCATAGTTCCTCGCACCTCTTCGGACATAGGAAAGAAATCTGGTTGGGATTAGACAAAGAGGGCGAA CCTTTCATGGCGATCAAGACCGTTCGGGAGATGCTCATGGTGGCCTATGATGTCCTAGAAAGTATGTCCATCTTT TCACGTCGATTGGGCAGATACTGACGGAACAAGTCACCCGCATGCTCTTCGCCCGACGCAGGGTCCTGCACCGCG ACATAAGCAAAGGGAATGTCATGTATATTTCCCGGCGCACAATGACACAGATCATTAAGACCATTAAGGCCCCCG ACGCGACTGCCGTAGGAAAGAATCAAGGAGACGGTTCCTGCTACTGTTTCGTCAGGTATTTGCTCGGCGAAAGGT AGGTGGCTGGTCTAGCTCTCTTCATATCTGACGACATGTTTCAGCATCGATTCTATGGAGACTTCCGCCCTGTTA ATCGATTTCAACTATAGTGAAATAGTGCCTGAGGACGAGTCAACAGGGGGACATGCGGTAGGTGCCCGAACAGTG AGATGCCATGTTTTCTTTCAGAAAGGTCCTGTCACTCACCTGTTGGATCAAGGGAACTCCCATGTTCATGGCCAG AGCCGTTCAAAATGGAGAGCCGATCAAGCTCGAGCCTTATCACACGGTCACCTTGGTCCCGATACCAAGCTCTGC ACACTTGGGTACCTACGAGAAAAACTTCCCGGACCGCATTTCTCGGTTCCGTGAGCTGGACAGGAAGGAGAATAT TGAGGCAAACGAAGTCGGGGAGCCCACGCACGAGATTCTCCTCCCCTGGAGACAACGACTGTATCATGATGCCGA ATCGGTCTATTGGCTCATGTACTATTGGGGCTTGCTGGCAAAGCCTTCGACCAGTATCAAGGAGAAAGGGGCGGA AGATTTCGACCCGGGCATCCCTGCGAACATCTGGACAGAATTGCACGCGGGCCGCAAGACGCTCTCTGCTCACGC GAACGACACATTCCATCCCTCCTACAGCCCTCTTCTTGATCTCTTCGTGCAAATGAGGGCCTACCTGCTGACTGA TCCTCATTGGTTCCCGCACGACTCTCCCAGGGCTGAGCCGGATTACATCCACGAATGTTTTCAGCGGCTTATTCT CAGATTTCTAAAGGACAACGTCGATAAGCCATTCATGGAATTGAAGAAGTCGGATCATGAGCGCCTCCCCTTCGC TGTTCCCGGGATCCCTGCTCGTTCCTCCTCGACAACTGATACCTACCTGTCGGATCAGCGAGCTCAAAGGCCTGA AACTCCTCCAATCCCCCGTGAGCCGTTCGCTGTCAATGTGCGTATCTCGCTTGTTTCTATATCCTTCTAGCATTA ACGTTGTTTCCCTTAGTATGTGGCACCCCCAGTCGTGGCTGGCTTCCAATCGACCGATAATACCCCGATTCAGCG CACGATGTCCCTCCCAGGCAAAGAGGGCGATGAAGACAGTGTCATTGAACAGCTTGTTTTCCCCCAAGGTTCGAG CCGCCAGGGTCCTGGTCGATCCCAAGAGTCGAACAACGATGATGTCTTCACTTGAGGTGCAGCTTCCCTCATTCT TTCTTCCCCTCTGAATTGCGCTTTCACTATGCTGTCATCGCTGCCGTTGTTACTTCATGTTGATGTGTTTTCCGG TTTCGATACCGATTTCCGTGTACAATATACGTAATGAGTATTTCAGCGTTGTCGA |
| Length | 2830 |