CopciAB_451158
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_451158 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 451158 |
| Uniprot id | Functional description | Cellulase (glycosyl hydrolase family 5) | |
| Location | scaffold_9:634631..637780 | Strand | + |
| Gene length (nt) | 3150 | Transcript length (nt) | 2521 |
| CDS length (nt) | 2316 | Protein length (aa) | 771 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7263297 | 64.3 | 0 | 1108 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB17369 | 63.8 | 0 | 1086 |
| Pleurotus eryngii ATCC 90797 | Pleery1_592183 | 64.1 | 0 | 1076 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1091243 | 64.1 | 0 | 1076 |
| Pleurotus ostreatus PC9 | PleosPC9_1_114491 | 64.1 | 0 | 1076 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_73767 | 65.1 | 0 | 1063 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_10427 | 62.5 | 0 | 1040 |
| Lentinula edodes NBRC 111202 | Lenedo1_736401 | 62.5 | 0 | 1039 |
| Flammulina velutipes | Flave_chr09AA00318 | 63 | 0 | 1013 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_75777 | 58.7 | 2.04E-304 | 987 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_194782 | 57.1 | 9.619E-304 | 985 |
| Grifola frondosa | Grifr_OBZ77054 | 58.2 | 1.059E-304 | 962 |
| Schizophyllum commune H4-8 | Schco3_2622489 | 61.7 | 6.438E-291 | 960 |
| Auricularia subglabra | Aurde3_1_1228860 | 52.1 | 1.556E-275 | 858 |
| Lentinula edodes B17 | Lened_B_1_1_11912 | 68.9 | 1.105E-134 | 447 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_451158.T0 |
| Description | Cellulase (glycosyl hydrolase family 5) |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 122 | 305 |
| Pfam | PF18564 | Glycoside hydrolase family 5 C-terminal domain | IPR041036 | 635 | 725 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| 1 | 743 | 765 | 22 |
InterPro
| Accession | Description |
|---|---|
| IPR017853 | Glycoside hydrolase superfamily |
| IPR001547 | Glycoside hydrolase, family 5 |
| IPR013780 | Glycosyl hydrolase, all-beta |
| IPR041036 | Glycoside hydrolase family 5, C-terminal domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| G | Cellulase (glycosyl hydrolase family 5) |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH5 | GH5_12 |
Transcription factor
| Group |
|---|
| No records |
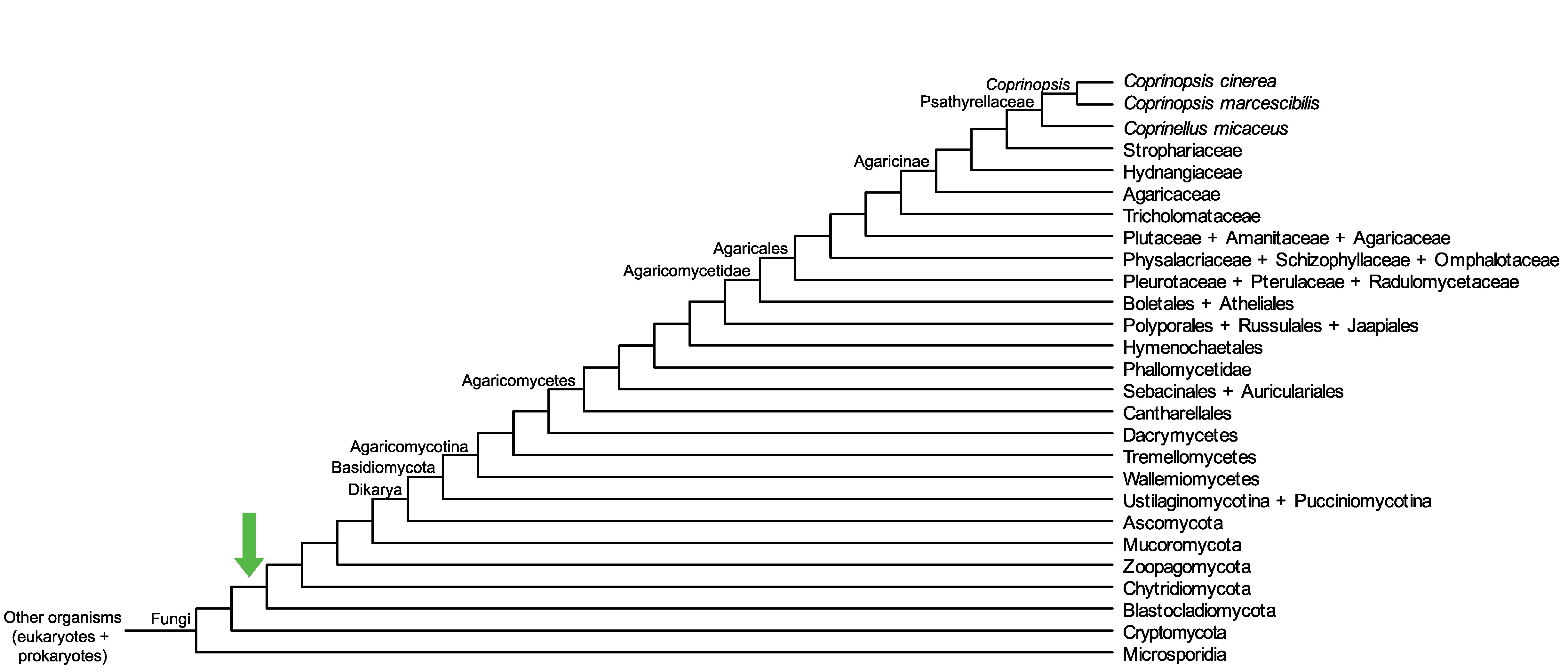
Conservation of CopciAB_451158 across fungi.
Arrow shows the origin of gene family containing CopciAB_451158.
Protein
| Sequence id | CopciAB_451158.T0 |
|---|---|
| Sequence |
>CopciAB_451158.T0 MSIPSTLDSSFILVENDARKPNPIVQDAKPNIPATHPSLEPGYKPGSANTYAHDWSPLATGGSIKTQGRHFVDGY GRVCHLRGANVSGCCKTPFDHDHENFPGDHRSVTFVGRPFPLEEAPEHFARLRRWGLTFIRFLVTWEAVEHAGPG IYDKEYLAYIRALLSLLPKYGLTAFVSMHQDVWSRYSGGSGAPAWTLEAAGFDLGAIEETGAGWLQGQKRGGHTE PDRGIWPCGYQKLSAATMATLFWGGDTFAPKLRVKGTDGEEVTIQKYLQDAFLGTWEEIVKAVGDLEGVIGVQMM NEPHRGYIELPSLHSFDYNTDLHLAHVPSAFQSFQLGAGHPTEVPHYERSFPWPTKLTGNTLVNANGVKAWREDG PTNGKCIWELHNVWGWDQKKNEAVILREGYFLVHPKTKEKVDWYVDCYFPLMQRWEKRIRPSIGADKMIFAEPIP NEFCPKTWTEERRPSNLVYAPHWYDLNALFSKAFGDFTVNVQGLSRGMFLPKALYWGQKGARDNYSLQIRNIVQK GYEALGDKPIVIGECGIPMDMNNKEAFETGDFTWHARMMDALMTALERSFVGFTLWNYNPSNNDVHGDDWNGENF SWFSRYRALPPSLLDYSQTSPTLDNGGRILTSIVRPYPAKTCGIPIKMEYEMTTGTFTYEWANSSGPLLSAETEI FYPSLIAQSRDVMVNGLEKGDRYVYDERRQTLFIVCQDVKTPGKRHRVEVSVSPGLDYPRPEFYLNDIWTDFGGQ ISAAVFVVLLVMALLTTRWLV |
| Length | 771 |
Coding
| Sequence id | CopciAB_451158.T0 |
|---|---|
| Sequence |
>CopciAB_451158.T0 ATGTCGATACCCTCAACATTAGACAGTTCCTTTATTCTCGTCGAAAACGACGCTCGCAAACCAAATCCTATCGTC CAGGATGCTAAACCCAATATTCCGGCAACGCACCCTTCGCTTGAACCAGGATATAAACCGGGTTCAGCAAATACA TACGCTCACGACTGGTCACCTTTAGCGACGGGAGGCTCAATAAAGACCCAAGGCCGCCATTTTGTCGACGGCTAC GGGCGAGTCTGCCATCTTCGCGGTGCCAACGTATCAGGCTGTTGCAAGACACCATTCGACCATGACCACGAAAAC TTCCCTGGGGACCATCGCTCTGTTACCTTTGTTGGCCGTCCCTTTCCCTTGGAGGAGGCTCCGGAGCACTTTGCA AGGCTGAGGAGATGGGGGTTAACTTTCATTCGTTTTCTTGTTACTTGGGAGGCCGTTGAGCACGCTGGACCAGGT ATCTACGACAAGGAATACCTTGCTTATATTCGAGCTCTTCTCTCCCTTCTCCCCAAGTACGGCCTGACCGCCTTC GTATCGATGCATCAAGATGTCTGGTCTCGTTACTCTGGTGGCTCAGGAGCGCCTGCCTGGACGCTGGAAGCTGCA GGATTCGACCTCGGCGCAATCGAAGAAACCGGAGCGGGTTGGCTTCAGGGTCAGAAGCGGGGCGGGCATACTGAA CCTGACCGCGGCATCTGGCCTTGCGGGTACCAGAAGCTGAGCGCAGCTACTATGGCAACGCTGTTCTGGGGCGGA GATACCTTCGCTCCAAAGCTCCGTGTCAAGGGGACGGATGGGGAGGAGGTCACCATCCAGAAGTACCTCCAAGAC GCCTTCCTTGGTACCTGGGAAGAAATAGTCAAGGCTGTTGGCGACCTGGAAGGCGTGATCGGCGTCCAAATGATG AATGAGCCTCATCGAGGCTACATCGAGCTACCTTCCCTTCATAGCTTTGACTACAACACAGATTTGCATCTTGCC CACGTCCCATCTGCTTTCCAATCGTTCCAGCTCGGCGCTGGTCACCCAACCGAAGTTCCTCACTACGAACGCTCC TTCCCCTGGCCGACCAAACTGACTGGAAACACCCTCGTCAACGCGAACGGCGTGAAAGCCTGGAGAGAAGACGGG CCTACCAACGGAAAGTGTATCTGGGAGCTGCACAACGTATGGGGATGGGACCAGAAGAAAAACGAAGCTGTTATC CTGAGGGAAGGATACTTCCTGGTACATCCGAAGACTAAAGAAAAGGTTGACTGGTATGTCGATTGCTATTTCCCT CTCATGCAGAGGTGGGAGAAGCGTATTCGACCGAGCATCGGTGCGGATAAGATGATCTTTGCCGAACCGATTCCG AACGAGTTCTGCCCCAAGACATGGACTGAGGAACGCCGTCCCTCAAATCTGGTGTATGCGCCCCACTGGTACGAC CTCAATGCTCTTTTCTCCAAGGCCTTCGGCGACTTTACTGTCAATGTACAAGGCCTCTCTCGGGGCATGTTCCTC CCGAAGGCCCTCTACTGGGGCCAGAAAGGTGCCAGGGATAACTACAGTCTTCAGATTCGCAACATTGTCCAGAAG GGTTATGAAGCCCTCGGGGACAAGCCTATTGTTATCGGCGAATGTGGTATTCCAATGGACATGAATAACAAAGAG GCTTTCGAGACTGGAGACTTTACCTGGCACGCTCGCATGATGGACGCGTTGATGACCGCCTTGGAACGGTCATTC GTTGGTTTTACCCTGTGGAATTACAATCCCTCAAACAACGATGTCCACGGCGACGACTGGAACGGCGAGAACTTT TCATGGTTCAGTCGTTATCGAGCCCTGCCACCCTCCCTGCTCGACTACTCCCAAACATCACCCACCTTGGATAAC GGTGGACGAATCCTCACCTCAATCGTCCGGCCGTACCCAGCCAAGACCTGCGGGATACCGATCAAGATGGAATAT GAAATGACGACGGGGACATTCACCTACGAATGGGCCAACTCCTCAGGGCCCCTGCTCTCCGCCGAGACAGAGATC TTCTACCCATCGCTTATCGCCCAGTCAAGGGACGTCATGGTTAATGGCCTGGAGAAAGGGGACCGATACGTCTAC GACGAGCGACGCCAGACTTTGTTCATCGTCTGCCAGGACGTCAAGACACCCGGCAAGCGACACCGAGTGGAGGTG TCTGTTTCACCTGGATTGGACTATCCTCGACCTGAGTTTTATCTTAACGATATTTGGACGGACTTTGGAGGGCAG ATTTCTGCTGCTGTTTTTGTTGTTTTGTTGGTTATGGCGTTGCTCACGACCCGGTGGTTGGTTTAA |
| Length | 2316 |
Transcript
| Sequence id | CopciAB_451158.T0 |
|---|---|
| Sequence |
>CopciAB_451158.T0 GGTGACGGGTTGATCCTGGCTGGCCATGTCGATACCCTCAACATTAGACAGTTCCTTTATTCTCGTCGAAAACGA CGCTCGCAAACCAAATCCTATCGTCCAGGATGCTAAACCCAATATTCCGGCAACGCACCCTTCGCTTGAACCAGG ATATAAACCGGGTTCAGCAAATACATACGCTCACGACTGGTCACCTTTAGCGACGGGAGGCTCAATAAAGACCCA AGGCCGCCATTTTGTCGACGGCTACGGGCGAGTCTGCCATCTTCGCGGTGCCAACGTATCAGGCTGTTGCAAGAC ACCATTCGACCATGACCACGAAAACTTCCCTGGGGACCATCGCTCTGTTACCTTTGTTGGCCGTCCCTTTCCCTT GGAGGAGGCTCCGGAGCACTTTGCAAGGCTGAGGAGATGGGGGTTAACTTTCATTCGTTTTCTTGTTACTTGGGA GGCCGTTGAGCACGCTGGACCAGGTATCTACGACAAGGAATACCTTGCTTATATTCGAGCTCTTCTCTCCCTTCT CCCCAAGTACGGCCTGACCGCCTTCGTATCGATGCATCAAGATGTCTGGTCTCGTTACTCTGGTGGCTCAGGAGC GCCTGCCTGGACGCTGGAAGCTGCAGGATTCGACCTCGGCGCAATCGAAGAAACCGGAGCGGGTTGGCTTCAGGG TCAGAAGCGGGGCGGGCATACTGAACCTGACCGCGGCATCTGGCCTTGCGGGTACCAGAAGCTGAGCGCAGCTAC TATGGCAACGCTGTTCTGGGGCGGAGATACCTTCGCTCCAAAGCTCCGTGTCAAGGGGACGGATGGGGAGGAGGT CACCATCCAGAAGTACCTCCAAGACGCCTTCCTTGGTACCTGGGAAGAAATAGTCAAGGCTGTTGGCGACCTGGA AGGCGTGATCGGCGTCCAAATGATGAATGAGCCTCATCGAGGCTACATCGAGCTACCTTCCCTTCATAGCTTTGA CTACAACACAGATTTGCATCTTGCCCACGTCCCATCTGCTTTCCAATCGTTCCAGCTCGGCGCTGGTCACCCAAC CGAAGTTCCTCACTACGAACGCTCCTTCCCCTGGCCGACCAAACTGACTGGAAACACCCTCGTCAACGCGAACGG CGTGAAAGCCTGGAGAGAAGACGGGCCTACCAACGGAAAGTGTATCTGGGAGCTGCACAACGTATGGGGATGGGA CCAGAAGAAAAACGAAGCTGTTATCCTGAGGGAAGGATACTTCCTGGTACATCCGAAGACTAAAGAAAAGGTTGA CTGGTATGTCGATTGCTATTTCCCTCTCATGCAGAGGTGGGAGAAGCGTATTCGACCGAGCATCGGTGCGGATAA GATGATCTTTGCCGAACCGATTCCGAACGAGTTCTGCCCCAAGACATGGACTGAGGAACGCCGTCCCTCAAATCT GGTGTATGCGCCCCACTGGTACGACCTCAATGCTCTTTTCTCCAAGGCCTTCGGCGACTTTACTGTCAATGTACA AGGCCTCTCTCGGGGCATGTTCCTCCCGAAGGCCCTCTACTGGGGCCAGAAAGGTGCCAGGGATAACTACAGTCT TCAGATTCGCAACATTGTCCAGAAGGGTTATGAAGCCCTCGGGGACAAGCCTATTGTTATCGGCGAATGTGGTAT TCCAATGGACATGAATAACAAAGAGGCTTTCGAGACTGGAGACTTTACCTGGCACGCTCGCATGATGGACGCGTT GATGACCGCCTTGGAACGGTCATTCGTTGGTTTTACCCTGTGGAATTACAATCCCTCAAACAACGATGTCCACGG CGACGACTGGAACGGCGAGAACTTTTCATGGTTCAGTCGTTATCGAGCCCTGCCACCCTCCCTGCTCGACTACTC CCAAACATCACCCACCTTGGATAACGGTGGACGAATCCTCACCTCAATCGTCCGGCCGTACCCAGCCAAGACCTG CGGGATACCGATCAAGATGGAATATGAAATGACGACGGGGACATTCACCTACGAATGGGCCAACTCCTCAGGGCC CCTGCTCTCCGCCGAGACAGAGATCTTCTACCCATCGCTTATCGCCCAGTCAAGGGACGTCATGGTTAATGGCCT GGAGAAAGGGGACCGATACGTCTACGACGAGCGACGCCAGACTTTGTTCATCGTCTGCCAGGACGTCAAGACACC CGGCAAGCGACACCGAGTGGAGGTGTCTGTTTCACCTGGATTGGACTATCCTCGACCTGAGTTTTATCTTAACGA TATTTGGACGGACTTTGGAGGGCAGATTTCTGCTGCTGTTTTTGTTGTTTTGTTGGTTATGGCGTTGCTCACGAC CCGGTGGTTGGTTTAATGTTGTGTGTTTTAAGTATGTAGATACCCTAATGTATAGAATCTAGAACTACATTGTTC ATTGCTGGATCCTCACACTTTCCTTGCATCGTTTTCATTTGTTGGGATTACTTATTTACGGGGATCAGTATTAAT AGAACGTCTATCAATAGTTGCCGTATTGCGGCTTGACCTCGTTTCA |
| Length | 2521 |
Gene
| Sequence id | CopciAB_451158.T0 |
|---|---|
| Sequence |
>CopciAB_451158.T0 GGTGACGGGTTGATCCTGGCTGGCCATGTCGATACCCTCAACATTAGACAGTTCCTTTATTCTCGTCGAAAACGA CGCTCGCAAACCAAATCCTATCGTCCAGGATGCTAAACCCAATATTCCGGTGAGTAGCATCATCCCAGAACTCTG AGCTCGAGCTAATTTAGCGCGCGACAGGCAACGCACCCTTCGCTTGAACCAGGATATAAACCGGGTTCAGCAAAT ACATACGCTCACGACTGGTCACCTTTAGCGACGGGAGGCTCAATAAAGACCCAAGGCCGCCATTTTGTCGACGGC TACGGGCGAGTCTGCCATCTTCGCGGTGCCAACGTATCAGGCTGTTGCAAGACGTCCGTTGTCTACAGCCTCCCG CATTACATCGTCTGACAACGCTTCTTTAGACCATTCGACCATGACCACGAAAACTTCCCTGGGGACCATCGCTCT GTTACCTTTGTTGGCCGTCCCTTTCCCTTGGAGGAGGCTCCGGAGCACTTTGCAAGGCTGAGGAGATGGGGGTTA ACTTTCAGTGAGTGTGGACGGGCTCTAGGACGCGTCTTTATTCACCAAGGTTCTCAGTTCGTTTTCTTGTTACTT GGGAGGCCGTTGAGCACGCTGGACCGTACGTCTCATTGAAGTGACCCGTCTCCGGTTGGCACAAACTGACTTTGC AGCTAAAGAGGTATCTACGACAAGGAATACCTTGCTTATATTCGAGCTCTTCTCTCCCTTCTCCCCAAGTACGGC CTGACCGCCTTCGTATCGATGCATCAAGATGTCTGGTCTCGTTACTCTGGTGGCTCAGGAGCGCCTGCCTGGACG CTGGAAGCTGCAGGATTCGACCTCGGCGCAATCGAAGAAACCGGAGCGGGTTGGCTTCAGGGTCAGAAGCGGGGC GGGCATACTGAACCTGACCGCGGCATCTGGCCTTGCGGGTACCAGAAGCTGAGCGCAGCTACTATGGCGTAAGTC TATTCCCTTTCTGCTCAGGTACATCGCATCGACTGAAACGCGTCGCTTAGAACGCTGTTCTGGGGCGGAGATACC TTCGCTCCAAAGCTCCGTGTCAAGGGGACGGATGGGGAGGAGGTCACCATCCAGAAGTACCTCCAAGACGCCTTC CTTGGTACCTGGGAAGAAATAGTCAAGGCTGTTGGCGACCTGGAAGGCGTGATCGGCGTCCAAGTATGTAGTGAC TACGTTTCCTGGTTTCAAAGCTTACCCCCTCTACAGATGATGAATGAGCCTCATCGAGGCTACATCGAGCTACCT TCCCTTCATAGCTTTGACTACAACACAGATTTGCATCTTGCCCACGTCCGTAAGTACGGACTTCGCCTTCAATTG AACTCGTACTAAAGTTTTGTATAGCATCTGCTTTCCAATCGTTCCAGCTCGGCGCTGGTCACCCAACCGAAGTTC CTCACTACGAACGCTCCTTCCCCTGGCCGACCAAACTGACTGGAAACACCCTCGTCAACGCGAACGGCGTGAAAG CCTGGAGAGAAGACGGGCCTACCAACGGAAAGTGTATCTGGGAGCTGCACAACGTATGGGGATGGGACCAGAAGA AAAACGAAGCTGTTATCCTGAGGGAAGGATACTTCCTGGTACATCCGAAGACTAAAGAAAAGGTTGACTGGTATG TCGATTGCTATTTCCCTCTCATGCAGAGGTGGGAGAAGCGTATTCGACCGAGCATCGGTGCGGATAAGATGATCT TTGCCGAACCGATTCCGAACGAGGTATGTACGATGCTCGGTAATAGGTTCTTATCTAATTGGCATGTGTTGTCAG TTCTGCCCCAAGACATGGACTGAGGAACGCCGTCCCTCAAATCTGGTGTATGCGCCCCACTGGTTAGTCCTCAGA TGCCTTCATTTATTTACCTTGCTGACATTGGATTGCTAGGTACGACCTCAATGCTCTTTTCTCCAAGGCCTTCGG CGACTTTACTGTCAATGTACAAGGCCTCTCTCGGGTAGGTCCTTCGCTTGACTCTTGAGTGTCTTGCCTATTTTC TTACATTCACCTAGGGCATGTTCCTCCCGAAGGCCCTCTACTGGGGCCAGAAAGGTGCCAGGGATAACTACAGTC TTCAGATTCGCAACATTGTCCAGAAGGGTTATGAAGCCCTCGGGGACAAGCCTATTGTTATCGGCGAATGTGGTA TTCCAATGGACATGAAGTGAGTGTCTGGATCTTAGAAACAATCCGCTGTTGCTGAACCGCTGCTGCAGTAACAAA GAGGCTTTCGAGACTGGAGACTTTACCTGGCACGCTCGCATGATGGACGCGTTGATGACCGCCTTGGAACGGTCA TTCGTTGGTTTTACGTGAGTCTGTCCAAAGTACACGCAGCATCTTTCTTGATGCCATGTTTCCAGCCTGTGGAAT TACAATCCCTCAAACAACGATGTCCACGGCGACGACTGGAACGGCGAGAACTTTTCATGGTTCAGTCGTTATCGA GCCCTGCCACCCTCCCTGCTCGACTACTCCCAAACATCACCCACCTTGGATAACGGTGGACGAATCCTCACCTCA ATCGTCCGGCCGTACCCAGCCAAGACCTGCGGGATACCGATCAAGATGGAATATGAAATGACGACGGGGACATTC ACCTACGAATGGGCCAACTCCTCAGGGCCCCTGCTCTCCGCCGAGACAGAGATCTTCTACCCATCGCTTATCGCC CAGTCAAGGGACGTCATGGTTAATGGCCTGGAGAAAGGGGACCGATACGTCTACGACGAGCGACGCCAGACTTTG TTCATCGTCTGCCAGGACGTCAAGACACCCGGCAAGCGACACCGAGTGGAGGTGTCTGTTTCACCTGGATTGGAC TATCCTCGACCTGAGTTTTATCTTAACGATATTTGGACGGACTTTGGAGGGCAGATTTCTGCTGCTGTTTTTGTT GTTTTGTTGGTTATGGCGTTGCTCACGACCCGGTGGTTGGTTTAATGTTGTGTGTTTTAAGTATGTAGATACCCT AATGTATAGAATCTAGAACTACATTGTTCATTGCTGGATCCTCACACTTTCCTTGCATCGTTTTCATTTGTTGGG ATTACTTATTTACGGGGATCAGTATTAATAGAACGTCTATCAATAGTTGCCGTATTGCGGCTTGACCTCGTTTCA |
| Length | 3150 |