CopciAB_459913
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_459913 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 459913 |
| Uniprot id | Functional description | GMC oxidoreductase | |
| Location | scaffold_3:1002626..1005523 | Strand | - |
| Gene length (nt) | 2898 | Transcript length (nt) | 2378 |
| CDS length (nt) | 2070 | Protein length (aa) | 689 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN8547_gmcA |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB26168 | 65.2 | 3.618E-258 | 803 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_121723 | 65.2 | 6.742E-255 | 793 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_183868 | 65.1 | 1.557E-253 | 789 |
| Agrocybe aegerita | Agrae_CAA7266836 | 54.5 | 1.229E-231 | 726 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1450716 | 57.3 | 3.621E-227 | 713 |
| Pleurotus ostreatus PC9 | PleosPC9_1_55933 | 57.7 | 3.538E-225 | 707 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1047763 | 57.6 | 1.77E-224 | 705 |
| Schizophyllum commune H4-8 | Schco3_2643012 | 56.4 | 3.779E-218 | 687 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_459913.T0 |
| Description | GMC oxidoreductase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | 112 | 423 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | 537 | 679 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal |
| IPR012132 | Glucose-methanol-choline oxidoreductase |
| IPR036188 | FAD/NAD(P)-binding domain superfamily |
| IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF |
| GO:0050660 | flavin adenine dinucleotide binding | MF |
KEGG
| KEGG Orthology |
|---|
| K00108 |
EggNOG
| COG category | Description |
|---|---|
| E | GMC oxidoreductase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA3 | AA3_2 |
Transcription factor
| Group |
|---|
| No records |
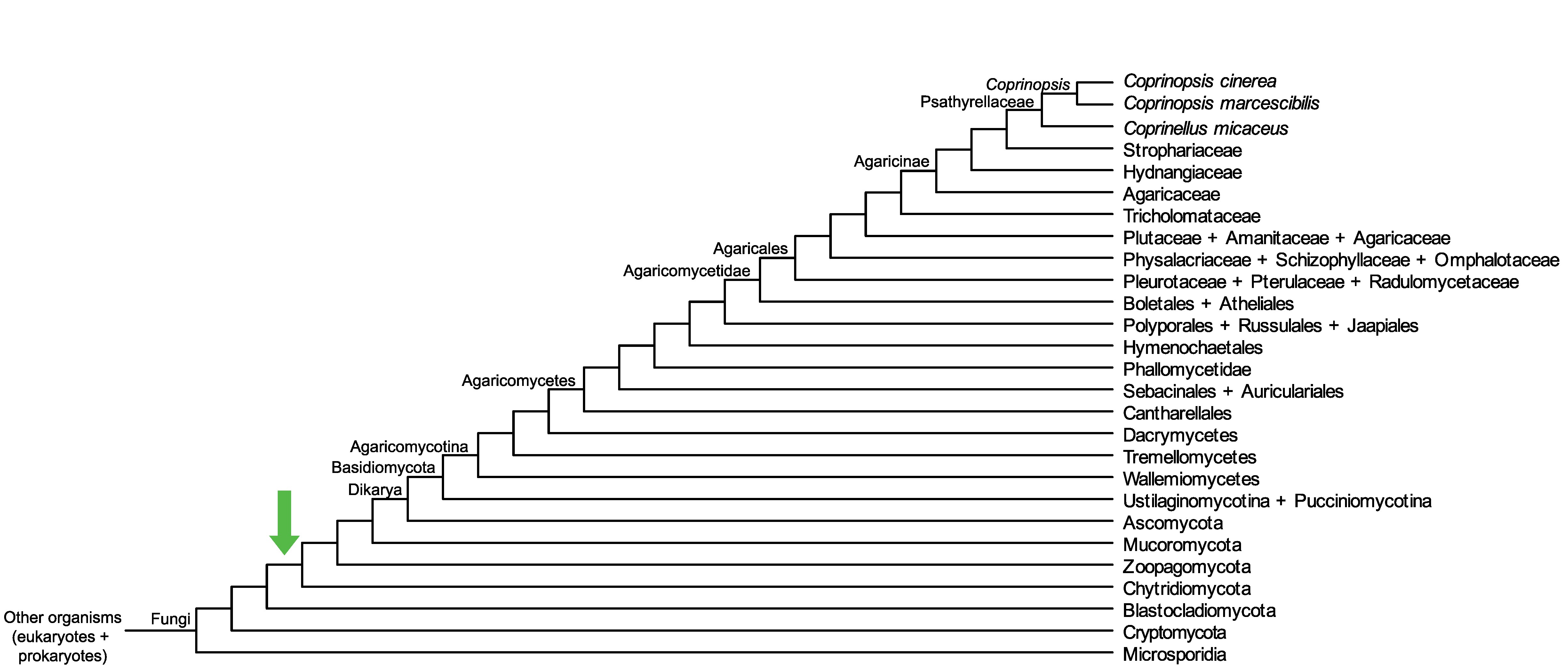
Conservation of CopciAB_459913 across fungi.
Arrow shows the origin of gene family containing CopciAB_459913.
Protein
| Sequence id | CopciAB_459913.T0 |
|---|---|
| Sequence |
>CopciAB_459913.T0 MASSNNALAPVPEALVELVTKLHEVVTTHPTGVPFIDALSPTTRALLASSSVLAVLVELLVRGQRVKSLKTKAES MNKAKGGSGQYVDSEDLGAAIEEARRLGGLAEKEWEYDVIVVGGGTSGCALAARLSEDPNLKVLLLEAGISGKTL PRSVMPSGFGGLFWGKHVHQLRTEPQQYAGGKTNFWPRAKMLGGCSSINAQMAQYGAPGDFDEWATVTGDDSWAW KNFAAYFRKFERFNPHPEYPDVDMKNKGTDGPVDVGYFNTVTPPSKAFIKACVAVGIPFTPDFNGLNGTLGVSRV MTYVSNQYRRVSSETAYLTPEVLGRKNLTVATNATATRIIFDTTSLGSAAEPRAVGVEFAKTEQGKRFKVYAKRD VVVSGGAVHTPHLLMLSGVGPQAHLEKLGIHVVKNHPNVGQNLVDHPVIDVYFKDKHNQSANYLKPKSLGDAVKL FKAIWQYKIEKTGGPLAMNFGESAAFVRSDDRSLFPADKFPEQLKDSTSAANSPDLEFFSTPFAYKEHGKIMFDV HTYALHCYLLRPTSKGEVLLKSANPFVQPSVNPNYLQTTEDVKKLARGLYLMLKIAQTEPLASHLDATFTREDLD HQTHLKSPQELEELVRERVETVYHPTTTCKMAPEDKGGVVDTKLRVYGIKGLRVCDASIFPYIISGHTAGGCFAI AEKLADEMKAEYRS |
| Length | 689 |
Coding
| Sequence id | CopciAB_459913.T0 |
|---|---|
| Sequence |
>CopciAB_459913.T0 ATGGCCTCTAGCAACAACGCACTCGCCCCTGTCCCTGAGGCCCTTGTCGAGCTAGTCACCAAGCTACACGAAGTC GTTACCACTCATCCTACCGGCGTCCCATTCATCGACGCGCTCTCGCCTACCACCCGTGCACTTCTCGCATCTAGC TCTGTCCTCGCAGTTCTCGTCGAACTCCTCGTCCGCGGTCAACGCGTCAAATCTTTGAAAACCAAAGCTGAGAGC ATGAACAAAGCGAAAGGAGGATCGGGCCAGTATGTCGACTCGGAGGATCTGGGAGCTGCGATAGAGGAGGCGAGG AGACTTGGAGGTCTTGCCGAGAAGGAATGGGAGTACGATGTCATTGTCGTTGGTGGAGGAACGAGTGGATGTGCA TTGGCTGCGCGCCTTTCCGAGGATCCGAATTTGAAGGTTCTTCTCCTCGAGGCTGGAATCTCTGGAAAGACCCTT CCTCGGTCCGTTATGCCCTCAGGCTTCGGTGGTCTGTTCTGGGGTAAACACGTTCACCAGTTGAGAACCGAGCCT CAGCAGTACGCTGGAGGAAAGACAAACTTCTGGCCTCGTGCAAAGATGCTCGGAGGATGCTCTTCGATCAATGCT CAAATGGCTCAATATGGCGCCCCAGGAGACTTTGACGAATGGGCCACAGTCACAGGGGATGACTCTTGGGCATGG AAGAACTTCGCCGCGTACTTTAGGAAGTTTGAGAGGTTCAATCCTCATCCGGAGTACCCCGACGTGGATATGAAG AACAAGGGCACTGACGGACCTGTCGACGTTGGCTACTTTAACACTGTAACCCCGCCTAGCAAGGCCTTTATCAAG GCTTGCGTTGCTGTTGGAATTCCTTTCACCCCGGATTTCAACGGCCTTAATGGAACGCTTGGAGTTAGCAGGGTA ATGACTTACGTTTCAAACCAATACAGGCGCGTATCGTCTGAGACAGCCTATCTCACCCCCGAGGTTCTCGGGCGC AAGAACCTTACTGTTGCCACCAATGCTACTGCCACCCGCATCATCTTTGACACCACCTCTCTCGGCTCAGCAGCC GAACCCAGGGCAGTCGGCGTTGAGTTTGCAAAGACCGAGCAAGGGAAGAGGTTCAAGGTCTATGCAAAGCGGGAC GTTGTCGTTTCCGGAGGAGCTGTCCACACCCCTCACCTCCTCATGCTCTCAGGTGTAGGACCTCAGGCCCACCTA GAGAAACTCGGTATCCACGTTGTGAAGAACCATCCCAACGTCGGTCAGAACCTTGTCGACCACCCTGTCATCGAC GTGTACTTCAAGGACAAGCATAACCAGAGTGCAAACTATTTGAAACCCAAGAGTCTCGGTGATGCCGTCAAGTTG TTCAAGGCTATCTGGCAGTACAAGATCGAGAAGACAGGCGGACCTTTGGCTATGAATTTTGGTGAATCGGCCGCG TTTGTGAGGTCGGATGATAGGAGCTTGTTCCCGGCGGACAAGTTCCCTGAGCAGTTGAAGGATAGCACTTCTGCC GCCAACAGCCCTGATCTCGAGTTCTTCTCCACGCCATTTGCATACAAGGAGCATGGAAAGATTATGTTTGATGTG CACACTTACGCTTTACACTGCTACCTGCTACGCCCCACGAGCAAAGGAGAAGTCCTCCTCAAGTCCGCCAACCCC TTCGTCCAGCCGAGCGTGAACCCCAACTACCTCCAAACCACCGAAGACGTGAAGAAACTCGCTCGGGGCCTCTAC CTCATGCTCAAGATCGCCCAAACCGAACCTCTCGCCTCGCATCTCGATGCTACCTTCACGCGTGAAGATCTCGAT CACCAAACTCACCTCAAGTCGCCTCAGGAATTGGAGGAACTCGTGAGGGAGCGTGTCGAGACCGTGTATCACCCC ACGACGACGTGCAAGATGGCACCAGAGGACAAGGGTGGTGTTGTTGATACCAAGTTGAGGGTGTATGGCATCAAG GGGTTGAGAGTGTGCGATGCTAGTATTTTCCCCTACATTATTTCTGGACACACGGCTGGTGGATGCTTTGCTATT GCCGAGAAGCTGGCAGATGAGATGAAGGCTGAGTATCGCTCCTAA |
| Length | 2070 |
Transcript
| Sequence id | CopciAB_459913.T0 |
|---|---|
| Sequence |
>CopciAB_459913.T0 CTCATTTGGGCGCTCTTTTCGACTTGTTTCACTGCGTCCAACGGTTACGCGGCACTTTTCCTTGCCTCTTCCTTG TCGAGCAGGGGATCTTTAGGCTTTTCTGACAAACATCAACGTCATCCCCGCTCATGGCCTCTAGCAACAACGCAC TCGCCCCTGTCCCTGAGGCCCTTGTCGAGCTAGTCACCAAGCTACACGAAGTCGTTACCACTCATCCTACCGGCG TCCCATTCATCGACGCGCTCTCGCCTACCACCCGTGCACTTCTCGCATCTAGCTCTGTCCTCGCAGTTCTCGTCG AACTCCTCGTCCGCGGTCAACGCGTCAAATCTTTGAAAACCAAAGCTGAGAGCATGAACAAAGCGAAAGGAGGAT CGGGCCAGTATGTCGACTCGGAGGATCTGGGAGCTGCGATAGAGGAGGCGAGGAGACTTGGAGGTCTTGCCGAGA AGGAATGGGAGTACGATGTCATTGTCGTTGGTGGAGGAACGAGTGGATGTGCATTGGCTGCGCGCCTTTCCGAGG ATCCGAATTTGAAGGTTCTTCTCCTCGAGGCTGGAATCTCTGGAAAGACCCTTCCTCGGTCCGTTATGCCCTCAG GCTTCGGTGGTCTGTTCTGGGGTAAACACGTTCACCAGTTGAGAACCGAGCCTCAGCAGTACGCTGGAGGAAAGA CAAACTTCTGGCCTCGTGCAAAGATGCTCGGAGGATGCTCTTCGATCAATGCTCAAATGGCTCAATATGGCGCCC CAGGAGACTTTGACGAATGGGCCACAGTCACAGGGGATGACTCTTGGGCATGGAAGAACTTCGCCGCGTACTTTA GGAAGTTTGAGAGGTTCAATCCTCATCCGGAGTACCCCGACGTGGATATGAAGAACAAGGGCACTGACGGACCTG TCGACGTTGGCTACTTTAACACTGTAACCCCGCCTAGCAAGGCCTTTATCAAGGCTTGCGTTGCTGTTGGAATTC CTTTCACCCCGGATTTCAACGGCCTTAATGGAACGCTTGGAGTTAGCAGGGTAATGACTTACGTTTCAAACCAAT ACAGGCGCGTATCGTCTGAGACAGCCTATCTCACCCCCGAGGTTCTCGGGCGCAAGAACCTTACTGTTGCCACCA ATGCTACTGCCACCCGCATCATCTTTGACACCACCTCTCTCGGCTCAGCAGCCGAACCCAGGGCAGTCGGCGTTG AGTTTGCAAAGACCGAGCAAGGGAAGAGGTTCAAGGTCTATGCAAAGCGGGACGTTGTCGTTTCCGGAGGAGCTG TCCACACCCCTCACCTCCTCATGCTCTCAGGTGTAGGACCTCAGGCCCACCTAGAGAAACTCGGTATCCACGTTG TGAAGAACCATCCCAACGTCGGTCAGAACCTTGTCGACCACCCTGTCATCGACGTGTACTTCAAGGACAAGCATA ACCAGAGTGCAAACTATTTGAAACCCAAGAGTCTCGGTGATGCCGTCAAGTTGTTCAAGGCTATCTGGCAGTACA AGATCGAGAAGACAGGCGGACCTTTGGCTATGAATTTTGGTGAATCGGCCGCGTTTGTGAGGTCGGATGATAGGA GCTTGTTCCCGGCGGACAAGTTCCCTGAGCAGTTGAAGGATAGCACTTCTGCCGCCAACAGCCCTGATCTCGAGT TCTTCTCCACGCCATTTGCATACAAGGAGCATGGAAAGATTATGTTTGATGTGCACACTTACGCTTTACACTGCT ACCTGCTACGCCCCACGAGCAAAGGAGAAGTCCTCCTCAAGTCCGCCAACCCCTTCGTCCAGCCGAGCGTGAACC CCAACTACCTCCAAACCACCGAAGACGTGAAGAAACTCGCTCGGGGCCTCTACCTCATGCTCAAGATCGCCCAAA CCGAACCTCTCGCCTCGCATCTCGATGCTACCTTCACGCGTGAAGATCTCGATCACCAAACTCACCTCAAGTCGC CTCAGGAATTGGAGGAACTCGTGAGGGAGCGTGTCGAGACCGTGTATCACCCCACGACGACGTGCAAGATGGCAC CAGAGGACAAGGGTGGTGTTGTTGATACCAAGTTGAGGGTGTATGGCATCAAGGGGTTGAGAGTGTGCGATGCTA GTATTTTCCCCTACATTATTTCTGGACACACGGCTGGTGGATGCTTTGCTATTGCCGAGAAGCTGGCAGATGAGA TGAAGGCTGAGTATCGCTCCTAAGGGTTTTGGGCTAGGGGATTTCTTGTGATTTATTCGCTTGTTTGTATCTACT GTACTCTCCTTGAACCTGTTGCATGTAATGATATCACTTCATTCTTTTCAGTTTGACTTATGTTACTGGCGCCTG TATGGACCCACAGGAATGAAACTGTATCCTGTTCTGATGCCCCGCTGGCTTTC |
| Length | 2378 |
Gene
| Sequence id | CopciAB_459913.T0 |
|---|---|
| Sequence |
>CopciAB_459913.T0 CTCATTTGGGCGCTCTTTTCGACTTGTTTCACTGCGTCCAACGGTTACGCGGCACTTTTCCTTGCCTCTTCCTTG TCGAGCAGGGGATCTTTAGGCTTTTCTGACAAACATCAACGTCGTGAGTAACGTATCTCAGTTCGAGCGGGTATA TGAACCACCAACGTGTCTTTTTCCAGATCCCCGCTCATGGCCTCTAGCAACAACGCACTCGCCCCTGTCCCTGAG GCCCTTGTCGAGCTAGTCACCAAGCTACACGAAGTCGTTACCACTCATCCTACCGGCGTCCCATTCATCGACGCG CTCTCGCCTACCACCCGTGCACTTCTCGCATCTAGCTCTGTCCTCGCAGTTCTCGTCGAACTCCTCGTCCGCGGT CAACGCGTCAAATCTTTGAAAACCAAAGCTGAGAGCATGAACAAAGCGAAAGGAGGATCGGGCCAGTATGTCGAC TCGGAGGATCTGGGAGCTGCGATAGAGGAGGCGAGGAGACTTGGAGGTCTTGCCGAGAAGGAATGGGAGTACGAT GTCATTGTCGTTGGTGGAGGAACGAGTGGATGTGCATTGGCTGCGCGCCTTTCCGAGGATCCGAATTTGAAGGTT CTTCTCCTCGAGGCTGGAATCTCGTATGTCTTTTTGATTCTGCCTGTAGGTTGTGCGCTGATGAGGTTTGAATAG TGGAAAGACCCTTCCTCGGTCCGTTATGCCCTCAGGCTTCGGTGGTCTGTTCTGGGGTAAACACGTTCACCAGTT GAGAACCGAGCCTCAGCAGTACGCTGGAGGAAAGACAAACTTCTGGCCTCGTGCAAAGATGCTCGGAGGATGCTC TTCGATCAATGCTCAAATGTACGTCTTCCACTCCCTCTTGTTCACCACTATGAATTGACCCTCTACTTAGGGCTC AATATGGCGCCCCAGGAGACTTTGACGAATGGGCCACAGTCACAGGGGATGACTCTTGGGCATGGAAGAACTTCG CCGCGTACTTTAGGAAGTTTGAGAGGTTCAATCCTCATCCGGAGTACCCCGACGTGGATATGAAGAACAAGGGCA CTGACGGACCTGTCGACGTTGGCTACTTTAACACTGTAACCCCGCCTAGCAAGGCCTTTATCAAGGCTTGCGTTG CTGTTGGAATTCCTTTCACCCCGGATTTCAACGGCCTTAATGGAACGCTTGGAGTTAGCAGGGTAAGTATCGACA CGTTCCCACCAAGAAAGTGCCCTAACCATTTCCATTCACGTTTCTCTGATAGATCAGTAAGGTTCTTCCATCACT TTCGAAAGACTTATTTTGACGCGTTTCATTACAGTGACTTACGTTTCAAACCAATACAGGCGCGTATCGTCTGAG ACAGCCTATCTCACCCCCGAGGTTCTCGGGCGCAAGAACCTTACTGTTGCCACCAATGCTACTGCCACCCGCATC ATCTTTGACACCACCTCTCTCGGCTCAGCAGCCGAACCCAGGGCAGTCGGCGTTGAGTTTGCAAAGACCGAGCAA GGGAAGAGGTTCAAGGTCTATGCAAAGCGGGACGTTGTCGTTTCGTGAGTCGCGCCTTGCCCTCCCACTTCCACT TTGCCCTGACCTGGACTTTGTAGCGGAGGAGCTGTCCACACCCCTCACCTCCTCATGCTCTCAGGTGTAGGACCT CAGGCCCACCTAGAGAAACTCGGTATCCACGTTGTGAAGAACCATCCCAACGTCGGTCAGAACCTTGTCGACCAC CCTGTCATCGACGTGTACTTCAAGGACAAGCATAACCAGAGTGCAAACTATTTGAAACCCAAGAGTCTCGGTGAT GCCGTCAAGTTGTTCAAGGCTATCTGGCAGTACAAGATCGAGAAGACAGGCGGACCTTTGGCTATGAATGTGAGC TTTTTCAATGGATGCCAACGAGGATTGTGGTTGTGCTGATACGGCGGTATCATAGTTTGGTGAATCGGCCGCGTT TGTGAGGTCGGATGATAGGAGCTTGTTCCCGGCGGACAAGTTCCCTGAGCAGTTGAAGGATAGCACTTCTGCCGC CAACAGCCCTGATCTCGAGTTCTTCTCCACGCCATTTGCATACAAGGAGCATGGAAAGATTATGTTTGATGTGCA CACTTACGCTTTACACTGCTACCTGCTACGGTACGTCCGATCCAGCTGGTCCTTTGGAACGGTTCTAATCTCTGG ATCTTCATTAGCCCCACGAGCAAAGGAGAAGTCCTCCTCAAGTCCGCCAACCCCTTCGTCCAGCCGAGCGTGAAC CCCAACTACCTCCAAACCACCGAAGACGTGAAGAAACTCGCTCGGGGCCTCTACCTCATGCTCAAGATCGCCCAA ACCGAACCTCTCGCCTCGCATCTCGATGCTACCTTCACGCGTGAAGATCTCGATCACCAAACTCACCTCAAGTCG CCTCAGGAATTGGAGGAACTCGTGAGGGAGCGTGTCGAGACCGTGTATCACCCCACGACGACGTGCAAGATGGCA CCAGAGGACAAGGGTGGTGTTGTTGATACCAAGTTGAGGGTGTATGGCATCAAGGGGTTGAGAGTGTGCGATGCT AGTATTTTCCCCTACATTATTTCTGGACACACGGTGAGTAGTTCTGTTGTCTGGCTGGAATCGTTGTGGGGATTA CTGATATGTTGTTTGTTCCAATTTTAGGCTGGTGGATGCTTTGCTATTGCCGAGAAGCTGGCAGATGAGATGAAG GCTGAGTATCGCTCCTAAGGGTTTTGGGCTAGGGGATTTCTTGTGATTTATTCGCTTGTTTGTATCTACTGTACT CTCCTTGAACCTGTTGCATGTAATGATATCACTTCATTCTTTTCAGTTTGACTTATGTTACTGGCGCCTGTATGG ACCCACAGGAATGAAACTGTATCCTGTTCTGATGCCCCGCTGGCTTTC |
| Length | 2898 |