CopciAB_462625
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_462625 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 462625 |
| Uniprot id | Functional description | Belongs to the glycosyl hydrolase 28 family | |
| Location | scaffold_10:129345..131609 | Strand | - |
| Gene length (nt) | 2265 | Transcript length (nt) | 2151 |
| CDS length (nt) | 1257 | Protein length (aa) | 418 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Neurospora crassa | gh28-2 |
| Aspergillus nidulans | AN10274_rhgB |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Auricularia subglabra | Aurde3_1_1309351 | 32.4 | 2.214E-66 | 233 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_3839 | 31.9 | 1.357E-66 | 233 |
| Lentinula edodes NBRC 111202 | Lenedo1_1220072 | 31.3 | 1.44E-65 | 230 |
| Flammulina velutipes | Flave_chr03AA00387 | 31.8 | 2.913E-65 | 229 |
| Lentinula edodes B17 | Lened_B_1_1_4785 | 32.4 | 1.448E-60 | 215 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_122672 | 33.7 | 1.831E-58 | 209 |
| Grifola frondosa | Grifr_OBZ69746 | 31.8 | 2.35E-51 | 188 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_462625.T0 |
| Description | Belongs to the glycosyl hydrolase 28 family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00295 | Glycosyl hydrolases family 28 | IPR000743 | 95 | 396 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 22 | 0.9975 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR012334 | Pectin lyase fold |
| IPR011050 | Pectin lyase fold/virulence factor |
| IPR000743 | Glycoside hydrolase, family 28 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004650 | polygalacturonase activity | MF |
| GO:0005975 | carbohydrate metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| G | Belongs to the glycosyl hydrolase 28 family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH28 |
Transcription factor
| Group |
|---|
| No records |
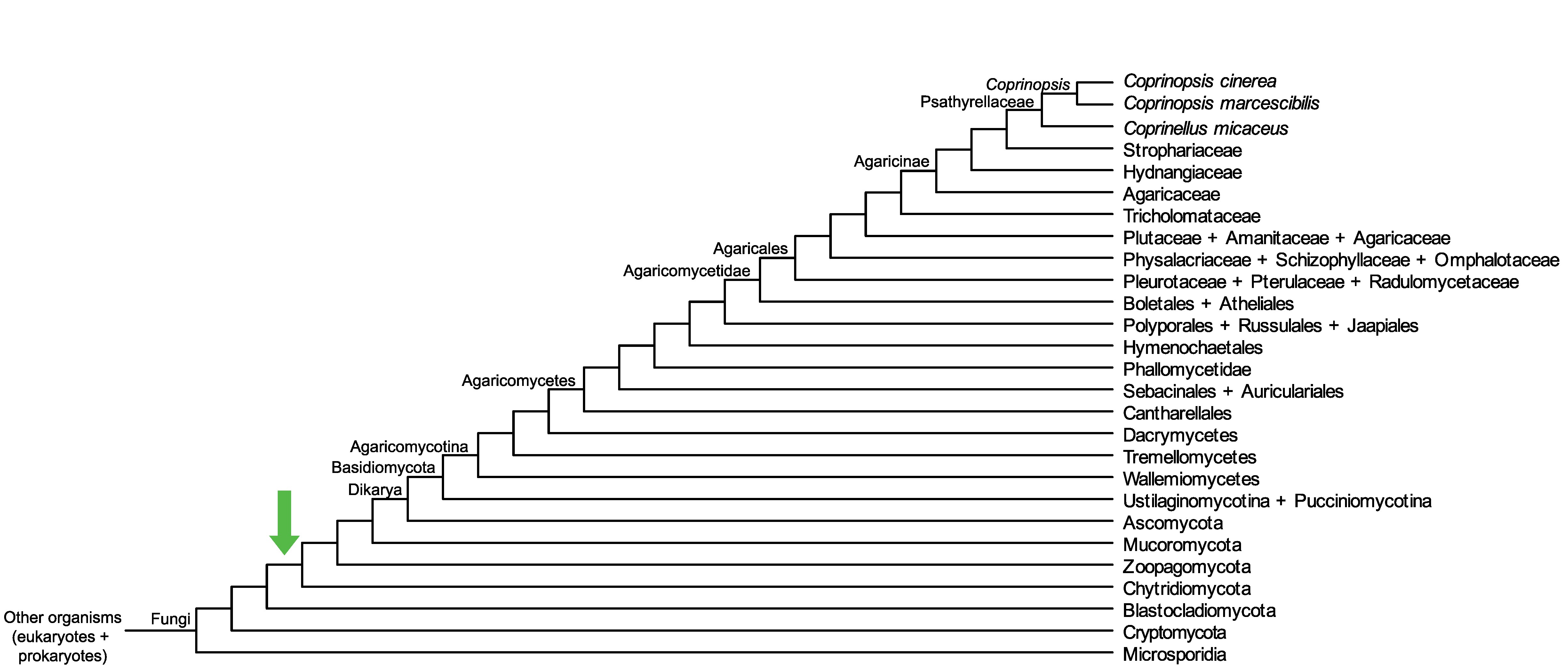
Conservation of CopciAB_462625 across fungi.
Arrow shows the origin of gene family containing CopciAB_462625.
Protein
| Sequence id | CopciAB_462625.T0 |
|---|---|
| Sequence |
>CopciAB_462625.T0 MKLAASFFASLCLLAGLHPVLAAVTQIGTACTVTPLSPTPVPPGGRPPDDTPQFRDAYSRCGRNGSITFTEGNFY IGQVMDVTLQNCEINILGTITWSTDIQYWLRNSIAVTYAQRHTAWRISGTNIAIRGHGKGLLFGNGQTWYDQNRN QGNQMGRPISLTIWRANNVFIDGLTWRQPQFWNTFVAYSQNVTMTNLDMEAKSNSQWTTVNTDGTNIWNSRDIYI SNWTVYSGDDCICAKGNTTNMHVKDITCYESGGMVVGSIGSVASQPDFVEHVLFENVRLNHSSNAAWIKTYPGQG YVNNVTFRNIQFSDVNQPIYITSCIYSGQNCDSSRLNIRNVRWENITGTSRYNVGAGMHCSASSPCQNLTFTGID IRQFNGGGPVKNFCSNIANQATMGLRCDGPCPGNWPQQLNGNR |
| Length | 418 |
Coding
| Sequence id | CopciAB_462625.T0 |
|---|---|
| Sequence |
>CopciAB_462625.T0 ATGAAGCTGGCCGCCTCGTTCTTCGCTAGTCTCTGCCTTTTGGCCGGGCTACACCCCGTCCTCGCTGCTGTTACT CAAATCGGGACTGCATGTACCGTCACACCCCTCTCCCCAACCCCCGTTCCGCCGGGCGGTCGTCCACCGGACGAC ACCCCTCAATTCCGCGACGCCTACAGCCGCTGCGGCCGTAACGGCTCCATCACGTTCACCGAAGGCAACTTCTAC ATCGGCCAAGTCATGGACGTCACCCTCCAAAACTGCGAAATCAACATCCTCGGCACCATCACCTGGAGCACCGAC ATTCAGTACTGGCTCCGAAACAGCATCGCGGTCACCTACGCCCAGAGGCACACGGCGTGGAGGATCAGTGGTACG AACATCGCGATTAGGGGCCATGGCAAGGGTTTATTGTTCGGAAATGGCCAGACGTGGTACGACCAGAATCGGAAC CAGGGTAACCAAATGGGAAGGCCTATTTCGCTCACCATTTGGAGGGCGAACAACGTCTTCATTGATGGGTTGACC TGGAGGCAGCCACAGTTCTGGAACACATTCGTTGCTTACTCGCAGAATGTGACCATGACCAACCTCGATATGGAG GCCAAGTCCAACTCTCAGTGGACCACGGTCAATACCGACGGCACCAACATCTGGAACTCTCGCGACATCTACATA TCTAACTGGACCGTCTACAGTGGCGACGATTGCATCTGCGCCAAAGGCAACACCACCAACATGCACGTCAAGGAC ATCACCTGTTACGAGTCTGGTGGAATGGTCGTGGGCAGCATCGGTTCCGTCGCCTCCCAACCCGACTTTGTCGAG CACGTCCTCTTCGAAAACGTGCGCCTCAACCACAGCAGCAATGCCGCCTGGATCAAGACCTACCCCGGCCAAGGC TACGTCAACAACGTCACCTTCCGCAACATCCAGTTCAGCGACGTGAACCAGCCTATCTACATCACCTCGTGCATC TACTCCGGCCAGAACTGCGATAGCTCGAGGTTGAATATCAGGAATGTGAGGTGGGAGAACATCACCGGGACGTCG AGGTATAACGTTGGCGCGGGGATGCATTGTAGTGCTTCGTCTCCGTGCCAGAACTTGACGTTTACGGGCATTGAT ATTAGGCAGTTCAATGGGGGAGGTCCGGTGAAGAACTTTTGCTCGAATATTGCGAACCAGGCGACGATGGGGCTC AGGTGCGATGGGCCGTGTCCGGGCAATTGGCCCCAGCAGTTGAACGGAAACAGATAG |
| Length | 1257 |
Transcript
| Sequence id | CopciAB_462625.T0 |
|---|---|
| Sequence |
>CopciAB_462625.T0 GCTCCTCCATGTCGACACTGAATTTGCTTTGGACTGACTTGGTCTCTACTCTATTTCTCTTCTTATGTCAAAAAA TTCCGTCGATTAAACTAACAATACCGGTTCATCAGGAAAATACGACTCGGGAACGCTCCAAAATCGCCAGCATCG TTCGACTGGAATGTTTCTTGGCTATCGCTTGGCTCTTCGTGACCGGCCTTGCTACTAGGAAGTCGCTTGGGCATT CTAATCTGTAGCCTTTTACCCATCTATCTAGCCGTAACCGTTCTCGCTGGCAAGCGGAAGAAACGGCCAGCGGAA CACGTTTTTCCGGAGGTCATTAGACCTCTTTTGGTGCTTCCCATCGCCAGCCCAATTTCATCGGATTCTCGAGAA AAAGACGATTTTGTCGTCGAAGGGCTAGGATATTGCTTCTCAACGGGCCGCTAATCTTGACTCTTTTCTTCTCTC GGGACGAGACCATTCCGACATTGGATACGAGACCCATTTCGGCCCTCCCAGCAGCCTTCTTTTCTACAACTGACG CTGTCTCATGATCTCCCCCTCTCCAGGCGCTCCAATGCACGGCGGTTCTTGAAGTATACAGACGGACCGCCAATC CCAACCTCGCAGGCTCCATGATTTTCATCGCATGAAAGCCTTCTTTTCTCTATCAGATTTAGAGTTGTCGCACTC GCATCATCCAAATCCGGAACGAAAATCATAACGAGGTTTAACCGTTTAGCGCTCTATTTAAAGTACAACCCCGAA CCACTCCTCTCTCCCAAGCTCGACTGAATCTCATCTCGAGGTTACAGAGTAATGAAGCTGGCCGCCTCGTTCTTC GCTAGTCTCTGCCTTTTGGCCGGGCTACACCCCGTCCTCGCTGCTGTTACTCAAATCGGGACTGCATGTACCGTC ACACCCCTCTCCCCAACCCCCGTTCCGCCGGGCGGTCGTCCACCGGACGACACCCCTCAATTCCGCGACGCCTAC AGCCGCTGCGGCCGTAACGGCTCCATCACGTTCACCGAAGGCAACTTCTACATCGGCCAAGTCATGGACGTCACC CTCCAAAACTGCGAAATCAACATCCTCGGCACCATCACCTGGAGCACCGACATTCAGTACTGGCTCCGAAACAGC ATCGCGGTCACCTACGCCCAGAGGCACACGGCGTGGAGGATCAGTGGTACGAACATCGCGATTAGGGGCCATGGC AAGGGTTTATTGTTCGGAAATGGCCAGACGTGGTACGACCAGAATCGGAACCAGGGTAACCAAATGGGAAGGCCT ATTTCGCTCACCATTTGGAGGGCGAACAACGTCTTCATTGATGGGTTGACCTGGAGGCAGCCACAGTTCTGGAAC ACATTCGTTGCTTACTCGCAGAATGTGACCATGACCAACCTCGATATGGAGGCCAAGTCCAACTCTCAGTGGACC ACGGTCAATACCGACGGCACCAACATCTGGAACTCTCGCGACATCTACATATCTAACTGGACCGTCTACAGTGGC GACGATTGCATCTGCGCCAAAGGCAACACCACCAACATGCACGTCAAGGACATCACCTGTTACGAGTCTGGTGGA ATGGTCGTGGGCAGCATCGGTTCCGTCGCCTCCCAACCCGACTTTGTCGAGCACGTCCTCTTCGAAAACGTGCGC CTCAACCACAGCAGCAATGCCGCCTGGATCAAGACCTACCCCGGCCAAGGCTACGTCAACAACGTCACCTTCCGC AACATCCAGTTCAGCGACGTGAACCAGCCTATCTACATCACCTCGTGCATCTACTCCGGCCAGAACTGCGATAGC TCGAGGTTGAATATCAGGAATGTGAGGTGGGAGAACATCACCGGGACGTCGAGGTATAACGTTGGCGCGGGGATG CATTGTAGTGCTTCGTCTCCGTGCCAGAACTTGACGTTTACGGGCATTGATATTAGGCAGTTCAATGGGGGAGGT CCGGTGAAGAACTTTTGCTCGAATATTGCGAACCAGGCGACGATGGGGCTCAGGTGCGATGGGCCGTGTCCGGGC AATTGGCCCCAGCAGTTGAACGGAAACAGATAGGATCGCTGTTGCCGGCTAGTACTTCGTAATGTACATTGGTCG GTTAGTCGCAAAGCGAGAATTTCTCGTCGAGGTTCTGAGGTGCGAGATGTC |
| Length | 2151 |
Gene
| Sequence id | CopciAB_462625.T0 |
|---|---|
| Sequence |
>CopciAB_462625.T0 GCTCCTCCATGTCGACACTGAATTTGCTTTGGACTGACTTGGTCTCTACTCTATTTCTCTTCTTATGTCAAAAAA TTCCGTCGATTAAACTAACAATACCGGTTCATCAGGAAAATACGACTCGGGAACGCTCCAAAATCGCCAGCATCG TTCGACTGGAATGTTTCTTGGCTATCGCTTGGCTCTTCGTGACCGGCCTTGCTACTAGGAAGTCGCTTGGGCATT CTAATCTGTAGCCTTTTACCCATCTATCTAGCCGTAACCGTTCTCGCTGGCAAGCGGAAGAAACGGCCAGCGGAA CACGTTTTTCCGGAGGTCATTAGACCTCTTTTGGTGCTTCCCATCGCCAGCCCAATTTCATCGGATTCTCGAGAA AAAGACGATTTTGTCGTCGAAGGGCTAGGATATTGCTTCTCAACGGGCCGCTAATCTTGACTCTTTTCTTCTCTC GGGACGAGACCATTCCGACATTGGATACGAGACCCATTTCGGCCCTCCCAGCAGCCTTCTTTTCTACAACTGACG CTGTCTCATGATCTCCCCCTCTCCAGGCGCTCCAATGCACGGCGGTTCTTGAAGTATACAGACGGACCGCCAATC CCAACCTCGCAGGCTCCATGATTTTCATCGCATGAAAGCCTTCTTTTCTCTATCAGATTTAGAGTTGTCGCACTC GCATCATCCAAATCCGGAACGAAAATCATAACGAGGTTTAACCGTTTAGCGCTCTATTTAAAGTACAACCCCGAA CCACTCCTCTCTCCCAAGCTCGACTGAATCTCATCTCGAGGTTACAGAGTAATGAAGCTGGCCGCCTCGTTCTTC GCTAGTCTCTGCCTTTTGGCCGGGCTACACCCCGTCCTCGCTGCTGTTACTCAAATCGGGACTGCATGTACCGTC ACACCCCTCTCCCCAACCCCCGTTCCGCCGGGCGGTCGTCCACCGGACGACACCCCTCAATTCCGCGACGCCTAC AGCCGCTGCGGCCGTAACGGCTCCATCACGTTCACCGAAGGCAACTTCTACATCGGCCAAGTCATGGACGTCACC CTCCAAAACTGCGAAATCAACATCCTCGGCACCATCACCTGGAGCACCGACATTCAGTACTGGCTCCGAAACAGC ATCGCGGTCACCTACGCCCAGAGGCACACGGCGTGGAGGATCAGTGGTACGAACATCGCGATTAGGGGCCATGGC AAGGGTTTATTGTTCGGAAATGGCCAGACGTGGTACGACCAGAATCGGAACCAGGGTAACCAAATGGGAAGGCCT ATTTCGCTCACCATTTGGAGGGCGAACAACGTCTTCATTGATGGGTTGACCTGGAGGCAGCCACAGTTCTGGTGA GTGGTTTCTTCAAGCGCGTTCCTGTGGGCTGGGCTCATGTTGAAAATTAGGAACACATTCGTTGCTTACTCGCAG AATGTGACCATGACCAACCTCGATATGGAGGCCAAGTCCAACTCTCAGTGGACCACGGTCAATACCGACGGCACC AACATCTGGAACTCTCGCGACATCTACATATCTAACTGGACCGTCTACAGTGGCGACGTAAGTCCTTCCCTTCAC GCCGTCCGATTGGCTCATTATTTCATTGCCTTCTGTGGGTAGGATTGCATCTGCGCCAAAGGCAACACCACCAAC ATGCACGTCAAGGACATCACCTGTTACGAGTCTGGTGGAATGGTCGTGGGCAGCATCGGTTCCGTCGCCTCCCAA CCCGACTTTGTCGAGCACGTCCTCTTCGAAAACGTGCGCCTCAACCACAGCAGCAATGCCGCCTGGATCAAGACC TACCCCGGCCAAGGCTACGTCAACAACGTCACCTTCCGCAACATCCAGTTCAGCGACGTGAACCAGCCTATCTAC ATCACCTCGTGCATCTACTCCGGCCAGAACTGCGATAGCTCGAGGTTGAATATCAGGAATGTGAGGTGGGAGAAC ATCACCGGGACGTCGAGGTATAACGTTGGCGCGGGGATGCATTGTAGTGCTTCGTCTCCGTGCCAGAACTTGACG TTTACGGGCATTGATATTAGGCAGTTCAATGGGGGAGGTCCGGTGAAGAACTTTTGCTCGAATATTGCGAACCAG GCGACGATGGGGCTCAGGTGCGATGGGCCGTGTCCGGGCAATTGGCCCCAGCAGTTGAACGGAAACAGATAGGAT CGCTGTTGCCGGCTAGTACTTCGTAATGTACATTGGTCGGTTAGTCGCAAAGCGAGAATTTCTCGTCGAGGTTCT GAGGTGCGAGATGTC |
| Length | 2265 |