CopciAB_478871
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_478871 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 478871 |
| Uniprot id | Functional description | Ste ste11 protein kinase | |
| Location | scaffold_1:2845583..2848645 | Strand | - |
| Gene length (nt) | 3063 | Transcript length (nt) | 2808 |
| CDS length (nt) | 2538 | Protein length (aa) | 845 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB29221 | 36.7 | 3.806E-130 | 437 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_1005 | 33.1 | 7.708E-127 | 427 |
| Lentinula edodes NBRC 111202 | Lenedo1_1124773 | 33.6 | 3.68E-126 | 425 |
| Agrocybe aegerita | Agrae_CAA7258667 | 34.3 | 9.126E-123 | 415 |
| Lentinula edodes B17 | Lened_B_1_1_18077 | 32.5 | 7.099E-122 | 412 |
| Schizophyllum commune H4-8 | Schco3_2684679 | 33.6 | 2.213E-121 | 411 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_103706 | 34 | 9.025E-108 | 370 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_140569 | 33.8 | 4.048E-107 | 368 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_478871.T0 |
| Description | Ste ste11 protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 95 | 359 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR017441 | Protein kinase, ATP binding site |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005524 | ATP binding | MF |
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K11229 |
EggNOG
| COG category | Description |
|---|---|
| T | Ste ste11 protein kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
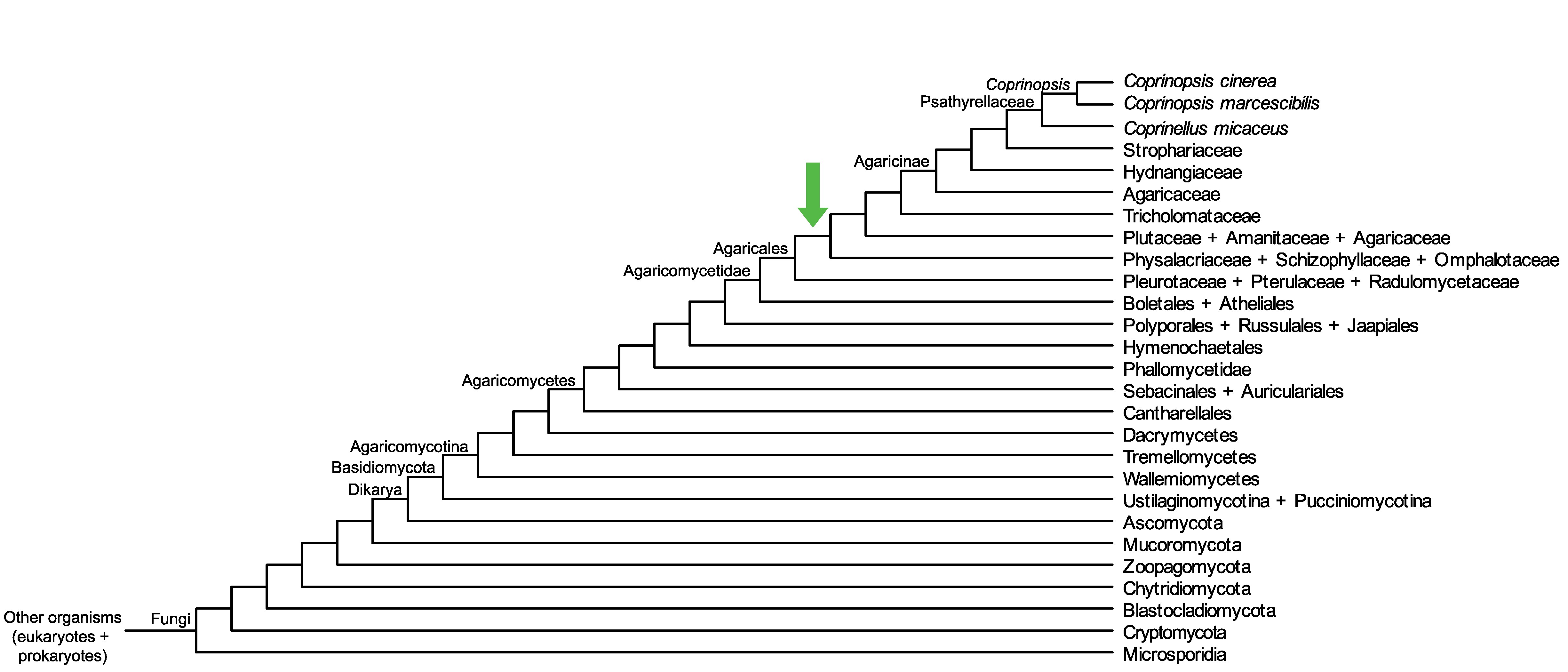
Conservation of CopciAB_478871 across fungi.
Arrow shows the origin of gene family containing CopciAB_478871.
Protein
| Sequence id | CopciAB_478871.T0 |
|---|---|
| Sequence |
>CopciAB_478871.T0 MDSRPASTLRTLQTWAVPFRETVPPVPDLPGRLLPQPSSSPHNSALDNAPDYSDSDDDDSIWQARPRERQPRRSS GQRLPYQRSFVKPTFQWIRGELLGKGSYGRVYLALNATTGEVMAVKQVELPKTPSDKMKPQQLDVMKALKFEGDT LKDLDHPNIVSYLGFEESQDYLSIFLEYVPGGTVGSLLVKNGRLREEVTKSWLRQILQGLDYLHGKGILHRDLKA DNILVDHSGVCKISDFGISKKAGEINRAKAHTGMKGTSFWMAPEILSPGEKGYDVKVDIWSVGCVAVEMWTGKRP WYGHEWWPVLMKVAEDKSPPPIPEDIPMGHLAQSFHNQCFRPDPADRPHASVMLKHPYLEIQSPWVFNWDDLADN SRPPRIYPELVPPVVRERTVTQSTFKANGPPTEKPRAHKPPLEPLKIPAYGPTSTQRKPSVNGAGPPVVWITPPN TPRRTAKPEDSSASTSTSDAAPWKSTSKRRLVIVNPDLDDDIGPAPQNTPPKPKFVYTPPPLPDPPQPPYASSPR PLRHAVSTSFASVSSMGSISSKAGSPSRPRALVDPGLSSTRTPPRPLPNPLSPPLGRSPAPLPVLPSPSKRRGSS RDRSHTVHTTFDASASKLYEEGNPTAKASVHNLPISSSGSRTIRRLPISPQQLNHGRPATTTTPSMPGSSTPRPL RPQKSVPDVSIFAFSRDAYHSRHHPSFKSKSNRPLYTSRGSFDSEPEDINGASDNDSVSSTSTTHTKHPQRGESL LAMRPRTEDVYKDLEAWFPNHDLDRPVDGADTPASGALEPRHHRVDVRKSIRMIAKEQNRRSHNDPASSRRRTML WNTHMEELSSDKRKDSMPRF |
| Length | 845 |
Coding
| Sequence id | CopciAB_478871.T0 |
|---|---|
| Sequence |
>CopciAB_478871.T0 ATGGATAGTAGGCCCGCGTCCACCCTGCGCACCCTCCAGACATGGGCTGTCCCTTTCAGAGAGACAGTCCCACCC GTTCCAGATCTTCCTGGTCGCCTACTGCCCCAGCCATCTTCGTCTCCTCACAATTCTGCCCTTGACAATGCCCCG GACTACTCGGACTCGGACGACGATGATAGTATCTGGCAAGCTAGACCTCGCGAACGCCAACCTCGGAGATCTTCA GGCCAGCGCCTGCCCTACCAGCGATCGTTTGTTAAGCCAACGTTCCAATGGATCAGAGGGGAGCTTCTTGGGAAA GGAAGCTATGGACGGGTTTACCTTGCCCTCAACGCAACCACAGGGGAGGTCATGGCAGTCAAGCAGGTCGAGCTA CCAAAGACCCCCAGTGACAAGATGAAGCCTCAGCAGCTAGACGTTATGAAAGCTCTCAAATTCGAGGGAGATACC CTCAAGGACTTGGATCATCCCAATATTGTCTCTTACCTCGGGTTCGAAGAGAGCCAGGACTACTTGAGCATTTTC CTGGAATATGTTCCAGGCGGTACAGTCGGGTCTCTCCTGGTAAAAAATGGTCGACTGCGGGAGGAAGTCACCAAG TCATGGCTTAGACAAATTCTTCAAGGGTTGGACTACCTCCACGGTAAAGGCATCCTTCACAGAGACTTGAAGGCG GATAACATCCTCGTCGACCATTCCGGGGTTTGCAAGATATCAGACTTTGGTATCTCCAAGAAGGCAGGGGAAATT AATCGTGCAAAAGCGCACACCGGAATGAAAGGTACCAGTTTCTGGATGGCACCAGAGATACTGTCACCCGGAGAG AAAGGGTACGATGTCAAGGTTGACATCTGGAGTGTGGGCTGCGTGGCTGTGGAAATGTGGACCGGTAAACGCCCA TGGTACGGCCACGAATGGTGGCCTGTGTTGATGAAGGTTGCTGAGGACAAGTCACCGCCCCCAATCCCTGAGGAT ATACCGATGGGTCATCTGGCACAGAGTTTCCACAATCAATGTTTTCGCCCAGATCCCGCCGACCGACCACATGCC TCTGTGATGCTTAAGCACCCTTACCTGGAAATTCAATCTCCCTGGGTGTTCAATTGGGACGATCTGGCCGACAAT TCGAGACCGCCACGGATCTATCCTGAGCTAGTCCCTCCAGTAGTCCGCGAGCGGACCGTAACCCAATCCACATTC AAAGCTAATGGCCCACCTACAGAGAAGCCCAGGGCTCACAAACCCCCACTGGAGCCCCTCAAAATTCCTGCGTAT GGACCCACATCAACCCAAAGAAAACCCAGCGTCAATGGGGCTGGACCTCCAGTCGTGTGGATCACCCCTCCGAAC ACCCCACGGAGAACAGCCAAGCCGGAAGACTCTAGCGCTTCCACAAGTACCTCTGACGCAGCCCCTTGGAAGAGC ACTTCTAAGCGTCGACTTGTCATCGTCAACCCCGACCTTGACGACGACATCGGACCCGCACCCCAGAATACCCCA CCGAAACCCAAGTTCGTGTACACACCGCCCCCTCTGCCGGACCCTCCTCAGCCTCCGTATGCTTCTTCACCCCGA CCCTTACGCCATGCCGTTAGCACGAGTTTTGCTTCTGTGTCGTCGATGGGATCGATTTCCTCGAAGGCGGGCTCA CCCTCTCGCCCCCGTGCCCTGGTCGACCCAGGACTTTCTTCCACGCGGACACCCCCAAGGCCACTCCCGAACCCC TTGTCTCCTCCCCTAGGTCGGTCCCCTGCACCCCTCCCCGTCTTGCCCTCCCCTTCAAAGCGAAGGGGATCCAGC CGCGACAGATCACACACGGTACATACTACCTTTGACGCCTCGGCCTCTAAATTATACGAGGAAGGTAACCCAACC GCCAAAGCTTCTGTTCACAACCTCCCGATCTCCTCTTCCGGCTCCCGCACCATCCGCCGCCTTCCCATCTCGCCT CAACAGTTGAACCACGGCAGACCTGCAACCACGACCACACCTTCGATGCCAGGCTCCTCGACCCCTCGGCCACTG CGACCACAGAAATCAGTCCCTGATGTATCCATCTTTGCTTTCAGCCGCGACGCTTATCATTCCCGGCATCATCCT TCGTTCAAGTCGAAGAGCAATCGTCCACTTTACACCTCACGCGGAAGCTTCGACTCGGAACCAGAAGATATCAAC GGGGCATCTGACAACGACAGTGTTTCTTCGACGTCTACGACTCATACCAAGCATCCGCAGCGAGGCGAATCACTG TTGGCGATGCGGCCACGGACGGAGGACGTGTATAAAGATTTAGAGGCTTGGTTCCCGAATCATGATCTCGACCGG CCAGTGGATGGCGCCGACACACCCGCCTCGGGAGCGCTGGAACCCAGGCATCACAGGGTGGACGTCAGGAAGAGT ATTCGAATGATCGCCAAGGAGCAAAACAGGCGCTCTCATAACGATCCAGCTTCTTCTCGTCGGCGAACGATGCTG TGGAACACACATATGGAAGAACTTTCGAGCGACAAGAGAAAGGACAGCATGCCGCGCTTTTAG |
| Length | 2538 |
Transcript
| Sequence id | CopciAB_478871.T0 |
|---|---|
| Sequence |
>CopciAB_478871.T0 GCTCTCTGGCGATCCATCTCCAGTCCACTGTGATTCCCTCCGGCTAACCAGCATCTCCTAATCCACCCTCTCCAG TTTCCTATACACTCTGTCTCACCCAGGGTCGCCCACCGCCTTCCCCGTTCTTCCCATCTGCGACGGATGGATAGT AGGCCCGCGTCCACCCTGCGCACCCTCCAGACATGGGCTGTCCCTTTCAGAGAGACAGTCCCACCCGTTCCAGAT CTTCCTGGTCGCCTACTGCCCCAGCCATCTTCGTCTCCTCACAATTCTGCCCTTGACAATGCCCCGGACTACTCG GACTCGGACGACGATGATAGTATCTGGCAAGCTAGACCTCGCGAACGCCAACCTCGGAGATCTTCAGGCCAGCGC CTGCCCTACCAGCGATCGTTTGTTAAGCCAACGTTCCAATGGATCAGAGGGGAGCTTCTTGGGAAAGGAAGCTAT GGACGGGTTTACCTTGCCCTCAACGCAACCACAGGGGAGGTCATGGCAGTCAAGCAGGTCGAGCTACCAAAGACC CCCAGTGACAAGATGAAGCCTCAGCAGCTAGACGTTATGAAAGCTCTCAAATTCGAGGGAGATACCCTCAAGGAC TTGGATCATCCCAATATTGTCTCTTACCTCGGGTTCGAAGAGAGCCAGGACTACTTGAGCATTTTCCTGGAATAT GTTCCAGGCGGTACAGTCGGGTCTCTCCTGGTAAAAAATGGTCGACTGCGGGAGGAAGTCACCAAGTCATGGCTT AGACAAATTCTTCAAGGGTTGGACTACCTCCACGGTAAAGGCATCCTTCACAGAGACTTGAAGGCGGATAACATC CTCGTCGACCATTCCGGGGTTTGCAAGATATCAGACTTTGGTATCTCCAAGAAGGCAGGGGAAATTAATCGTGCA AAAGCGCACACCGGAATGAAAGGTACCAGTTTCTGGATGGCACCAGAGATACTGTCACCCGGAGAGAAAGGGTAC GATGTCAAGGTTGACATCTGGAGTGTGGGCTGCGTGGCTGTGGAAATGTGGACCGGTAAACGCCCATGGTACGGC CACGAATGGTGGCCTGTGTTGATGAAGGTTGCTGAGGACAAGTCACCGCCCCCAATCCCTGAGGATATACCGATG GGTCATCTGGCACAGAGTTTCCACAATCAATGTTTTCGCCCAGATCCCGCCGACCGACCACATGCCTCTGTGATG CTTAAGCACCCTTACCTGGAAATTCAATCTCCCTGGGTGTTCAATTGGGACGATCTGGCCGACAATTCGAGACCG CCACGGATCTATCCTGAGCTAGTCCCTCCAGTAGTCCGCGAGCGGACCGTAACCCAATCCACATTCAAAGCTAAT GGCCCACCTACAGAGAAGCCCAGGGCTCACAAACCCCCACTGGAGCCCCTCAAAATTCCTGCGTATGGACCCACA TCAACCCAAAGAAAACCCAGCGTCAATGGGGCTGGACCTCCAGTCGTGTGGATCACCCCTCCGAACACCCCACGG AGAACAGCCAAGCCGGAAGACTCTAGCGCTTCCACAAGTACCTCTGACGCAGCCCCTTGGAAGAGCACTTCTAAG CGTCGACTTGTCATCGTCAACCCCGACCTTGACGACGACATCGGACCCGCACCCCAGAATACCCCACCGAAACCC AAGTTCGTGTACACACCGCCCCCTCTGCCGGACCCTCCTCAGCCTCCGTATGCTTCTTCACCCCGACCCTTACGC CATGCCGTTAGCACGAGTTTTGCTTCTGTGTCGTCGATGGGATCGATTTCCTCGAAGGCGGGCTCACCCTCTCGC CCCCGTGCCCTGGTCGACCCAGGACTTTCTTCCACGCGGACACCCCCAAGGCCACTCCCGAACCCCTTGTCTCCT CCCCTAGGTCGGTCCCCTGCACCCCTCCCCGTCTTGCCCTCCCCTTCAAAGCGAAGGGGATCCAGCCGCGACAGA TCACACACGGTACATACTACCTTTGACGCCTCGGCCTCTAAATTATACGAGGAAGGTAACCCAACCGCCAAAGCT TCTGTTCACAACCTCCCGATCTCCTCTTCCGGCTCCCGCACCATCCGCCGCCTTCCCATCTCGCCTCAACAGTTG AACCACGGCAGACCTGCAACCACGACCACACCTTCGATGCCAGGCTCCTCGACCCCTCGGCCACTGCGACCACAG AAATCAGTCCCTGATGTATCCATCTTTGCTTTCAGCCGCGACGCTTATCATTCCCGGCATCATCCTTCGTTCAAG TCGAAGAGCAATCGTCCACTTTACACCTCACGCGGAAGCTTCGACTCGGAACCAGAAGATATCAACGGGGCATCT GACAACGACAGTGTTTCTTCGACGTCTACGACTCATACCAAGCATCCGCAGCGAGGCGAATCACTGTTGGCGATG CGGCCACGGACGGAGGACGTGTATAAAGATTTAGAGGCTTGGTTCCCGAATCATGATCTCGACCGGCCAGTGGAT GGCGCCGACACACCCGCCTCGGGAGCGCTGGAACCCAGGCATCACAGGGTGGACGTCAGGAAGAGTATTCGAATG ATCGCCAAGGAGCAAAACAGGCGCTCTCATAACGATCCAGCTTCTTCTCGTCGGCGAACGATGCTGTGGAACACA CATATGGAAGAACTTTCGAGCGACAAGAGAAAGGACAGCATGCCGCGCTTTTAGACAAAGCAATGTAAAATTTGT TAGTCGTAGTCATAGTGGAATCCTTCGCGCGCATATCATCTATACGGACACTTTTCTATATGGACTTTGTACTGT TGTACCAATAAGTAACCTTTTTCCATGACAAGG |
| Length | 2808 |
Gene
| Sequence id | CopciAB_478871.T0 |
|---|---|
| Sequence |
>CopciAB_478871.T0 GCTCTCTGGCGATCCATCTCCAGTCCACTGTGATTCCCTCCGGCTAACCAGCATCTCCTAATCCACCCTCTCCAG TTTCCTATACACTCTGTCTCACCCAGGGTCGCCCACCGCCTTCCCCGTTCTTCCCATCTGCGACGGATGGATAGT AGGCCCGCGTCCACCCTGCGCACCCTCCAGACATGGGCTGTCCCTTTCAGAGAGACAGTCCCACCCGTTCCAGAT CTTCCTGGTCGCCTACTGCCCCAGCCATCTTCGTCTCCTCACAATTCTGCCCTTGACAATGCCCCGGACTACTCG GACTCGGACGACGATGATAGTATCTGGCAAGCTAGACCTCGCGAACGCCAACCTCGGAGATCTTCAGGCCAGCGC CTGCCCTACCAGCGATCGTTTGTTAAGCGTATGTCTTACTCTTTTCTCTGAACATTCAACAGCTGACACGGATCC ATAAGCAACGTTCCAATGGATCAGAGGGGAGCTTCTTGGGAAAGGAAGCTATGGACGGGTTTACCTTGCCCTCAA CGCAACCACAGGGGAGGTCATGGCAGTCAAGCAGGTCGAGCTACCAAAGACCCCCAGTGACAAGATGAAGCCTCA GCAGCTAGACGTTATGAAAGCTCTCAAATTCGAGGGAGATACCCTCAAGGACTTGGATCATCCCAATATTGTCTC TTACCTCGGGTTCGAAGAGAGCCAGGACTACTTGAGCATGTAAGTGGTAACCTGCTGCTATGGAATCCCTGCTGA GAGCGGACAGTTTCCTGGAATATGTTCCAGGCGGTACAGTCGGGTCTCTCCTGGTAAAAAATGGTCGACTGCGGG AGGAAGTCACCAAGTCATGGCTTAGACAAATTCTTCAAGGGTTGGACTACCTCCACGGTAAAGGCATCCTTCACA GAGTCAGTTCCTCTTTTCGGTCTAACAGAGTTGTAATAACCGAATCGTCTCTCAGGACTTGAAGGCGGATAACAT CCTCGTCGACCATTCCGGGGTTTGCAAGATATCAGACTTTGGTATCTCCAAGAAGGCAGGGGAAATTAATCGTGC AAAAGCGCACACCGGAATGAAAGGTACCAGTTTCTGGATGGCACCAGAGATACTGTCACCCGGAGAGAAAGGGTA CGATGTCAAGGTTGACATCTGGAGTGTGGGCTGCGTGGCTGTGGAAATGTGGACCGGTAAACGCCCATGGTACGG CCACGAATGGTGGCCTGTGTTGATGAAGGTAACGGGAGACTCGTCTCTGTTGGCGCCATTCACTGACCTGTGCAC CACAGGTTGCTGAGGACAAGTCACCGCCCCCAATCCCTGAGGATATACCGATGGGTCATCTGGCACAGAGTTTCC ACAATCAATGTTTTCGCCCGTGAGTAATGTGTCGAAGTCCACCTCGTTCTTCTTCTCACAAGCTCGGGCAGAGAT CCCGCCGACCGACCACATGCCTCTGTGATGCTTAAGCACCCTTACCTGGAAATTCAATCTCCCTGGGTGTTCAAT TGGGACGATCTGGCCGACAATTCGAGACCGCCACGGATCTATCCTGAGCTAGTCCCTCCAGTAGTCCGCGAGCGG ACCGTAACCCAATCCACATTCAAAGCTAATGGCCCACCTACAGAGAAGCCCAGGGCTCACAAACCCCCACTGGAG CCCCTCAAAATTCCTGCGTATGGACCCACATCAACCCAAAGAAAACCCAGCGTCAATGGGGCTGGACCTCCAGTC GTGTGGATCACCCCTCCGAACACCCCACGGAGAACAGCCAAGCCGGAAGACTCTAGCGCTTCCACAAGTACCTCT GACGCAGCCCCTTGGAAGAGCACTTCTAAGCGTCGACTTGTCATCGTCAACCCCGACCTTGACGACGACATCGGA CCCGCACCCCAGAATACCCCACCGAAACCCAAGTTCGTGTACACACCGCCCCCTCTGCCGGACCCTCCTCAGCCT CCGTATGCTTCTTCACCCCGACCCTTACGCCATGCCGTTAGCACGAGTTTTGCTTCTGTGTCGTCGATGGGATCG ATTTCCTCGAAGGCGGGCTCACCCTCTCGCCCCCGTGCCCTGGTCGACCCAGGACTTTCTTCCACGCGGACACCC CCAAGGCCACTCCCGAACCCCTTGTCTCCTCCCCTAGGTCGGTCCCCTGCACCCCTCCCCGTCTTGCCCTCCCCT TCAAAGCGAAGGGGATCCAGCCGCGACAGATCACACACGGTACATACTACCTTTGACGCCTCGGCCTCTAAATTA TACGAGGAAGGTAACCCAACCGCCAAAGCTTCTGTTCACAACCTCCCGATCTCCTCTTCCGGCTCCCGCACCATC CGCCGCCTTCCCATCTCGCCTCAACAGTTGAACCACGGCAGACCTGCAACCACGACCACACCTTCGATGCCAGGC TCCTCGACCCCTCGGCCACTGCGACCACAGAAATCAGTCCCTGATGTATCCATCTTTGCTTTCAGCCGCGACGCT TATCATTCCCGGCATCATCCTTCGTTCAAGTCGAAGAGCAATCGTCCACTTTACACCTCACGCGGAAGCTTCGAC TCGGAACCAGAAGATATCAACGGGGCATCTGACAACGACAGTGTTTCTTCGACGTCTACGACTCATACCAAGCAT CCGCAGCGAGGCGAATCACTGTTGGCGATGCGGCCACGGACGGAGGACGTGTATAAAGATTTAGAGGCTTGGTTC CCGAATCATGATCTCGACCGGCCAGTGGATGGCGCCGACACACCCGCCTCGGGAGCGCTGGAACCCAGGCATCAC AGGGTGGACGTCAGGAAGAGTATTCGAATGATCGCCAAGGAGCAAAACAGGCGCTCTCATAACGATCCAGCTTCT TCTCGTCGGCGAACGATGCTGTGGAACACACATATGGAAGAACTTTCGAGCGACAAGAGAAAGGACAGCATGCCG CGCTTTTAGACAAAGCAATGTAAAATTTGTTAGTCGTAGTCATAGTGGAATCCTTCGCGCGCATATCATCTATAC GGACACTTTTCTATATGGACTTTGTACTGTTGTACCAATAAGTAACCTTTTTCCATGACAAGG |
| Length | 3063 |