CopciAB_485043
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_485043 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 485043 |
| Uniprot id | Functional description | Glycosyl hydrolase family 85 | |
| Location | scaffold_2:832509..835562 | Strand | + |
| Gene length (nt) | 3054 | Transcript length (nt) | 2777 |
| CDS length (nt) | 2430 | Protein length (aa) | 809 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Grifola frondosa | Grifr_OBZ65487 | 48.7 | 4.088E-151 | 497 |
| Auricularia subglabra | Aurde3_1_1413065 | 36.2 | 3.852E-101 | 350 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_485043.T0 |
| Description | Glycosyl hydrolase family 85 |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF03644 | Glycosyl hydrolase family 85 | IPR005201 | 75 | 409 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR005201 | Glycoside hydrolase, family 85 |
| IPR032979 | Cytosolic endo-beta-N-acetylglucosaminidase |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005737 | cytoplasm | CC |
| GO:0033925 | mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K01227 |
EggNOG
| COG category | Description |
|---|---|
| G | Glycosyl hydrolase family 85 |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH85 |
Transcription factor
| Group |
|---|
| No records |
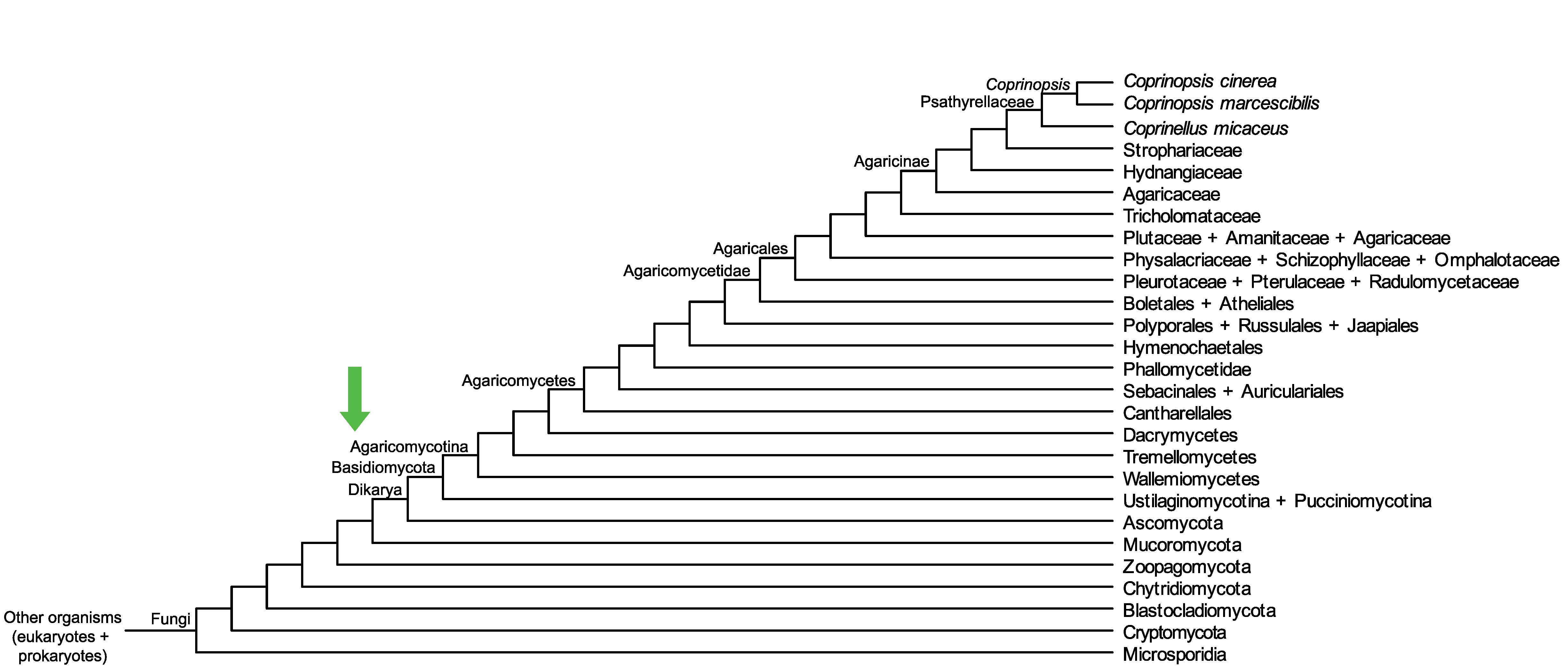
Conservation of CopciAB_485043 across fungi.
Arrow shows the origin of gene family containing CopciAB_485043.
Protein
| Sequence id | CopciAB_485043.T0 |
|---|---|
| Sequence |
>CopciAB_485043.T0 MPVRGTVPQPRALPEYFQSFQELDDWRTSKTGPFYSAEGVRPYVPRRLRTEDITGQGRLLIAHDFKGGYIEDPFT KSYTFNWWSSTSLFNYFAHHRITIPPPEWIASAHRQGVPILGTLMFEGEAQEDCFRMVIGTQPRNATVDDAPWSR ANYTVPVSPHYAEILADIAKERGFDGWLINIEMDLKGGSSQARGIAAWVSLFQQEVLKKVGSHGLVLWYDSVTFR GHLSWQERLSSRNLPFFLSSSGLFTNYAWYNHFPQRQIDYFNSLDPKLLKGKSLQDIYAGIDMWGRGSHGGGGFG MYKALEHADPQKLGLSIALLAPGWTWESENENPGWTWERFWEQDTNLWVGPSSSKVAIEVPPTEFKVVEPMCEHG PFKPVSKFFPTLPPPDPLDLAFYTSFSPGVGHQWFVEGKEVFYSGSGWTDIEKQTSTGDLVWPKPTVYNLVGGGL SDADASSALYFEDAWNGGNSLQINVTRTRGSTGYAGYWVPIQSLSLSTGNRYEASVVYKVGEGATGDVDAKLDLG VRGGGTRRNAGFKLLEEDTTALASGWAKARIVFEPSAAEGEESIVPASIGFIVSVSSDSTKSSPSNNSFLLPFLV GQIAVHPHLPADYHSFEASLYSLSFKRNATSRSPLDGMLSWDIVAALPEPPEPDESPDRDDPNLFWEVQPTERNW FPKFLYFNIYASEIADGNTSPSKENQKFWIGTTGFDGETRKFHVYDQNLPPSLQTKNGWIRRKGKKLTFEVEGVL ETGEIVRWGNQSRLYRPLLHDLEGSCRFSWFEPCSNSDLTSLLMWSTVLVVLWVKYKFY |
| Length | 809 |
Coding
| Sequence id | CopciAB_485043.T0 |
|---|---|
| Sequence |
>CopciAB_485043.T0 ATGCCAGTCCGCGGAACGGTACCCCAACCTAGAGCTCTTCCCGAATATTTCCAATCGTTCCAGGAGCTAGATGAC TGGAGAACGAGCAAGACAGGGCCGTTCTATTCTGCGGAGGGTGTTCGACCTTATGTTCCGAGGAGGTTGAGGACT GAGGATATTACTGGGCAAGGGAGGCTACTGATCGCGCATGACTTCAAGGGAGGTTATATTGAAGACCCATTCACA AAATCATATACTTTCAATTGGTGGTCATCTACCTCCCTATTCAATTACTTCGCACACCACCGGATAACCATTCCT CCACCAGAATGGATTGCCTCGGCACATAGACAGGGAGTCCCAATACTAGGAACACTGATGTTCGAAGGAGAAGCC CAGGAAGACTGCTTTCGCATGGTAATAGGAACGCAACCCCGCAACGCCACAGTGGACGACGCACCATGGTCGAGG GCCAATTACACCGTCCCAGTCTCCCCGCACTACGCGGAAATCCTCGCAGACATTGCCAAAGAACGCGGATTCGAC GGTTGGCTAATTAACATAGAAATGGATTTGAAGGGAGGGTCTTCGCAAGCTCGTGGAATAGCCGCTTGGGTATCT CTATTTCAGCAAGAGGTTTTGAAGAAGGTCGGGAGCCATGGCTTAGTGCTTTGGTACGACAGTGTCACGTTCAGA GGCCATCTTTCCTGGCAAGAGCGACTATCCTCTCGCAACCTGCCCTTCTTCCTCAGCTCGAGTGGGTTGTTCACC AACTACGCGTGGTACAACCATTTCCCCCAGCGCCAGATTGACTACTTCAATAGCCTCGACCCGAAACTCCTCAAG GGTAAATCCCTCCAAGACATTTACGCAGGTATCGACATGTGGGGACGAGGCTCGCACGGCGGCGGCGGATTCGGA ATGTACAAAGCCCTCGAGCACGCCGACCCTCAAAAGCTCGGACTCAGCATCGCACTCTTAGCTCCAGGCTGGACA TGGGAATCCGAAAACGAAAACCCGGGCTGGACATGGGAGCGTTTCTGGGAACAAGACACGAATTTATGGGTCGGA CCGTCCTCGAGTAAAGTGGCTATCGAAGTCCCGCCAACCGAGTTCAAGGTTGTCGAACCGATGTGTGAGCATGGG CCGTTTAAACCTGTGTCCAAATTCTTCCCGACTCTCCCCCCTCCGGATCCCCTGGATTTGGCATTTTATACGTCA TTTTCGCCGGGCGTTGGGCATCAGTGGTTTGTAGAGGGGAAGGAAGTCTTTTATTCAGGGTCTGGGTGGACGGAT ATCGAGAAGCAAACAAGTACGGGGGATTTGGTGTGGCCGAAGCCTACAGTGTATAACTTGGTGGGAGGAGGGCTG AGCGACGCGGATGCAAGCTCAGCGCTTTATTTCGAAGACGCTTGGAATGGCGGGAACTCGTTGCAGATTAATGTT ACCCGCACTAGGGGCTCGACGGGGTATGCAGGGTATTGGGTACCCATTCAGTCGCTTAGCCTGTCGACGGGGAAC CGCTATGAAGCCTCGGTGGTCTACAAAGTAGGAGAAGGGGCAACTGGGGATGTGGATGCGAAGCTGGATTTGGGA GTGAGAGGTGGTGGTACTAGGAGAAACGCAGGATTCAAGCTCTTGGAGGAGGATACGACTGCTTTGGCTTCTGGA TGGGCGAAGGCTCGGATTGTCTTCGAGCCATCTGCTGCTGAAGGCGAGGAAAGCATCGTGCCAGCGTCCATCGGC TTCATCGTATCCGTGTCCAGCGACTCCACGAAGTCCAGCCCAAGCAACAACTCATTCCTTCTCCCCTTCCTCGTC GGCCAGATTGCTGTACATCCACACCTCCCAGCCGACTACCATTCCTTCGAAGCATCGCTTTACTCGCTCAGCTTT AAACGCAACGCGACCTCTCGCAGTCCACTGGATGGCATGTTATCTTGGGATATCGTCGCAGCGCTCCCCGAACCT CCCGAACCAGACGAGTCTCCAGATCGAGATGACCCTAACCTTTTCTGGGAAGTTCAACCCACCGAGAGGAATTGG TTCCCCAAGTTCCTATACTTCAATATCTACGCATCTGAAATCGCCGACGGAAACACCAGCCCATCGAAGGAGAAC CAGAAATTCTGGATAGGCACCACAGGATTCGACGGTGAAACCCGCAAATTCCACGTCTACGACCAGAACCTTCCA CCCAGTTTGCAGACCAAGAACGGATGGATAAGGCGAAAGGGCAAGAAACTCACGTTTGAAGTGGAGGGTGTGTTG GAAACGGGAGAGATTGTGCGTTGGGGGAACCAATCGAGGCTGTATAGGCCTCTTTTGCATGATTTGGAGGGAAGT TGTAGGTTCTCGTGGTTTGAACCGTGCTCGAATTCAGACTTGACTTCGCTGCTTATGTGGAGCACTGTATTGGTC GTGTTGTGGGTCAAATACAAGTTCTACTGA |
| Length | 2430 |
Transcript
| Sequence id | CopciAB_485043.T0 |
|---|---|
| Sequence |
>CopciAB_485043.T0 ATGTATCTTTGCACCACCGTCTCTGCAGCCTCGACGACAGGGATCTTTACAAGTATCAAATCCGCCGTTTATATT CGTCTTCGAAGACCCTGTATAGTCGACAAACATGCGGTGATTCCCACCTCTCAAATCAGCTGAGGACCCAAGGAA TACACCCAATCCTCCGAGCTTTATGCGTTGAGAACTCGAATTGCTGGGAGACTTCGTTCTTGACAATCATGGGCC ATGGGACACGACACCGACCACGACTTTGACCTTCTCATATTACCTGCCCTTTAACCTATGCCAGTCCGCGGAACG GTACCCCAACCTAGAGCTCTTCCCGAATATTTCCAATCGTTCCAGGAGCTAGATGACTGGAGAACGAGCAAGACA GGGCCGTTCTATTCTGCGGAGGGTGTTCGACCTTATGTTCCGAGGAGGTTGAGGACTGAGGATATTACTGGGCAA GGGAGGCTACTGATCGCGCATGACTTCAAGGGAGGTTATATTGAAGACCCATTCACAAAATCATATACTTTCAAT TGGTGGTCATCTACCTCCCTATTCAATTACTTCGCACACCACCGGATAACCATTCCTCCACCAGAATGGATTGCC TCGGCACATAGACAGGGAGTCCCAATACTAGGAACACTGATGTTCGAAGGAGAAGCCCAGGAAGACTGCTTTCGC ATGGTAATAGGAACGCAACCCCGCAACGCCACAGTGGACGACGCACCATGGTCGAGGGCCAATTACACCGTCCCA GTCTCCCCGCACTACGCGGAAATCCTCGCAGACATTGCCAAAGAACGCGGATTCGACGGTTGGCTAATTAACATA GAAATGGATTTGAAGGGAGGGTCTTCGCAAGCTCGTGGAATAGCCGCTTGGGTATCTCTATTTCAGCAAGAGGTT TTGAAGAAGGTCGGGAGCCATGGCTTAGTGCTTTGGTACGACAGTGTCACGTTCAGAGGCCATCTTTCCTGGCAA GAGCGACTATCCTCTCGCAACCTGCCCTTCTTCCTCAGCTCGAGTGGGTTGTTCACCAACTACGCGTGGTACAAC CATTTCCCCCAGCGCCAGATTGACTACTTCAATAGCCTCGACCCGAAACTCCTCAAGGGTAAATCCCTCCAAGAC ATTTACGCAGGTATCGACATGTGGGGACGAGGCTCGCACGGCGGCGGCGGATTCGGAATGTACAAAGCCCTCGAG CACGCCGACCCTCAAAAGCTCGGACTCAGCATCGCACTCTTAGCTCCAGGCTGGACATGGGAATCCGAAAACGAA AACCCGGGCTGGACATGGGAGCGTTTCTGGGAACAAGACACGAATTTATGGGTCGGACCGTCCTCGAGTAAAGTG GCTATCGAAGTCCCGCCAACCGAGTTCAAGGTTGTCGAACCGATGTGTGAGCATGGGCCGTTTAAACCTGTGTCC AAATTCTTCCCGACTCTCCCCCCTCCGGATCCCCTGGATTTGGCATTTTATACGTCATTTTCGCCGGGCGTTGGG CATCAGTGGTTTGTAGAGGGGAAGGAAGTCTTTTATTCAGGGTCTGGGTGGACGGATATCGAGAAGCAAACAAGT ACGGGGGATTTGGTGTGGCCGAAGCCTACAGTGTATAACTTGGTGGGAGGAGGGCTGAGCGACGCGGATGCAAGC TCAGCGCTTTATTTCGAAGACGCTTGGAATGGCGGGAACTCGTTGCAGATTAATGTTACCCGCACTAGGGGCTCG ACGGGGTATGCAGGGTATTGGGTACCCATTCAGTCGCTTAGCCTGTCGACGGGGAACCGCTATGAAGCCTCGGTG GTCTACAAAGTAGGAGAAGGGGCAACTGGGGATGTGGATGCGAAGCTGGATTTGGGAGTGAGAGGTGGTGGTACT AGGAGAAACGCAGGATTCAAGCTCTTGGAGGAGGATACGACTGCTTTGGCTTCTGGATGGGCGAAGGCTCGGATT GTCTTCGAGCCATCTGCTGCTGAAGGCGAGGAAAGCATCGTGCCAGCGTCCATCGGCTTCATCGTATCCGTGTCC AGCGACTCCACGAAGTCCAGCCCAAGCAACAACTCATTCCTTCTCCCCTTCCTCGTCGGCCAGATTGCTGTACAT CCACACCTCCCAGCCGACTACCATTCCTTCGAAGCATCGCTTTACTCGCTCAGCTTTAAACGCAACGCGACCTCT CGCAGTCCACTGGATGGCATGTTATCTTGGGATATCGTCGCAGCGCTCCCCGAACCTCCCGAACCAGACGAGTCT CCAGATCGAGATGACCCTAACCTTTTCTGGGAAGTTCAACCCACCGAGAGGAATTGGTTCCCCAAGTTCCTATAC TTCAATATCTACGCATCTGAAATCGCCGACGGAAACACCAGCCCATCGAAGGAGAACCAGAAATTCTGGATAGGC ACCACAGGATTCGACGGTGAAACCCGCAAATTCCACGTCTACGACCAGAACCTTCCACCCAGTTTGCAGACCAAG AACGGATGGATAAGGCGAAAGGGCAAGAAACTCACGTTTGAAGTGGAGGGTGTGTTGGAAACGGGAGAGATTGTG CGTTGGGGGAACCAATCGAGGCTGTATAGGCCTCTTTTGCATGATTTGGAGGGAAGTTGTAGGTTCTCGTGGTTT GAACCGTGCTCGAATTCAGACTTGACTTCGCTGCTTATGTGGAGCACTGTATTGGTCGTGTTGTGGGTCAAATAC AAGTTCTACTGATCCGTGTCAGGTGTATGTTTACATTTGGTTAGTGGACTGCTAATGGTACATGGTTAGAGGTTT GG |
| Length | 2777 |
Gene
| Sequence id | CopciAB_485043.T0 |
|---|---|
| Sequence |
>CopciAB_485043.T0 ATGTATCTTTGCACCACCGTCTCTGCAGCCTCGACGACAGGGATCTTTACAAGTATCAAATCCGCCGTTTATATT CGTCTTCGAAGACCCTGTATAGTCGACAAACATGCGGTGATTCCCACCTCTCAAATCAGCTGAGGACCCAAGGAA TACACCCAATCCTCCGAGCTTTATGCGTTGAGAACTCGAATTGCTGGGAGACTTCGTTCTTGACAATCATGGGCC ATGGGACACGACACCGACCACGACTTTGACCTTCTCATATTACCTGCCCTTTAACCTATGCCAGTCCGCGGAACG GTACCCCAACCTAGAGCTCTTCCCGAATATTTCCAATCGTTCCAGGAGCTAGATGACTGGAGAACGAGCAAGACA GGGCCGTTCTATTCTGCGGAGGGTGTTCGACCTTATGTTCCGAGGAGGTTGAGGACTGAGGATATTACTGGGCAA GGGAGGCTACTGGTGAGTACACGAACCATAATCACTCCAGGCCATCTTTTGAGTTCGGTTCTTCTTCTAGATCGC GCATGACTTCAAGGTGAATACCAACGAACGACGTCTATGTCCCAGCATCGCCGTTCTGACTGGCCCAGGGAGGTT ATATTGAAGACCCATTCACAAAATCATATACTTTCAATTGGTGGTCATCTACCTCCCTATTCAATTAGTCGGTCT TCCTTTCGAACCCGTTCCCCCCAATTGCGTTAACACCCCCGTTTATAGCTTCGCACACCACCGGATAACCATTCC TCCACCAGAATGGATTGCCTCGGCACATAGACAGGGAGTCCCAATACTAGGAACACTGTGAGCAACCTCTCTTTC GAGAGGACTTCATGTACTCAAATGGAACTACGGCAGGATGTTCGAAGGAGAAGCCCAGGAAGACTGCTTTCGCAT GGTAATAGGAACGCAACCCCGCAACGCCACAGTGGACGACGCACCATGGTCGAGGGCCAATTACACCGTCCCAGT CTCCCCGCACTACGCGGAAATCCTCGCAGACATTGCCAAAGAACGCGGATTCGACGGTTGGCTAATTAACATAGA AATGGATTTGAAGGGAGGGTCTTCGCAAGCTCGTGGAATAGCCGCTTGGGTATCTCTATTTCAGCAAGAGGTTTT GAAGAAGGTCGGGAGCCATGGCTTAGTGCTTTGGTACGACAGTGTCACGTTCAGAGGCCATCTTTCCTGGCAAGA GCGACTATCCTCTCGCAACCTGCCCTTCTTCCTCAGCTCGAGTGGGTTGTTCACCAACTACGCGGTGCGTTCTCT TCTGTCTTCGGAAACTAGAATAGCCGAGCTGATACATGTACAGTGGTACAACCATTTCCCCCAGCGCCAGATTGA CTACTTCAATAGCCTCGACCCGAAACTCCTCAAGGGTAAATCCCTCCAAGACATTTACGCAGGTATCGACATGTG GGGACGAGGCTCGCACGGCGGCGGCGGATTCGGAATGTACAAAGCCCTCGAGCACGCCGACCCTCAAAAGCTCGG ACTCAGCATCGCACTCTTAGCTCCAGGCTGGACATGGGAATCCGAAAACGAAAACCCGGGCTGGACATGGGAGCG TTTCTGGGAACAAGACACGAATTTATGGGTCGGACCGTCCTCGAGTAAAGTGGCTATCGAAGTCCCGCCAACCGA GTTCAAGGTTGTCGAACCGATGTGTGAGCATGGGCCGTTTAAACCTGTGTCCAAATTCTTCCCGACTCTCCCCCC TCCGGATCCCCTGGATTTGGCATTTTATACGTCATTTTCGCCGGGCGTTGGGCATCAGTGGTTTGTAGAGGGGAA GGAAGTCTTTTATTCAGGGTCTGGGTGGACGGATATCGAGAAGCAAACAAGTACGGGGGATTTGGTGTGGCCGAA GCCTACAGTGTATAACTTGGTGGGAGGAGGGCTGAGCGACGCGGATGCAAGCTCAGCGCTTTATTTCGAAGACGC TTGGAATGGCGGGAACTCGTTGCAGATTAATGTTACCCGCACTAGGGGCTCGACGGGGTATGCAGGGTATTGGGT ACCCATTCAGTCGCTTAGCCTGTCGACGGGGAACCGCTATGAAGCCTCGGTGGTCTACAAAGTAGGAGAAGGGGC AACTGGGGATGTGGATGCGAAGCTGGATTTGGGAGTGAGAGGTGGTGGTACTAGGAGAAACGCAGGATTCAAGCT CTTGGAGGAGGATACGACTGCTTTGGCTTCTGGATGGGCGAAGGCTCGGATTGTCTTCGAGCCATCTGCTGCTGA AGGCGAGGAAAGCATCGTGCCAGCGTCCATCGGCTTCATCGTATCCGTGTCCAGCGACTCCACGAAGTCCAGCCC AAGCAACAACTCATTCCTTCTCCCCTTCCTCGTCGGCCAGATTGCTGTACATCCACACCTCCCAGCCGACTACCA TTCCTTCGAAGCATCGCTTTACTCGCTCAGCTTTAAACGCAACGCGACCTCTCGCAGTCCACTGGATGGCATGTT ATCTTGGGATATCGTCGCAGCGCTCCCCGAACCTCCCGAACCAGACGAGTCTCCAGATCGAGATGACCCTAACCT TTTCTGGGAAGTTCAACCCACCGAGAGGAATTGGTTCCCCAAGTTCCTATACTTCAATATCTACGCATCTGAAAT CGCCGACGGAAACACCAGCCCATCGAAGGAGAACCAGAAATTCTGGATAGGCACCACAGGATTCGACGGTGAAAC CCGCAAATTCCACGTCTACGACCAGAACCTTCCACCCAGTTTGCAGACCAAGAACGGATGGATAAGGCGAAAGGG CAAGAAACTCACGTTTGAAGTGGAGGGTGTGTTGGAAACGGGAGAGATTGTGCGTTGGGGGAACCAATCGAGGCT GTATAGGCCTCTTTTGCATGATTTGGAGGGAAGTTGTAGGTTCTCGTGGTTTGAACCGTGCTCGAATTCAGACTT GACTTCGCTGCTTATGTGGAGCACTGTATTGGTCGTGTTGTGGGTCAAATACAAGTTCTACTGATCCGTGTCAGG TGTATGTTTACATTTGGTTAGTGGACTGCTAATGGTACATGGTTAGAGGTTTGG |
| Length | 3054 |