CopciAB_492628
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_492628 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 492628 |
| Uniprot id | Functional description | Belongs to the glycosyl hydrolase 5 (cellulase A) family | |
| Location | scaffold_2:1668285..1670748 | Strand | + |
| Gene length (nt) | 2464 | Transcript length (nt) | 1974 |
| CDS length (nt) | 1710 | Protein length (aa) | 569 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Pleurotus ostreatus PC15 | PleosPC15_2_1060958 | 55.3 | 1.618E-213 | 667 |
| Pleurotus ostreatus PC9 | PleosPC9_1_75659 | 55.3 | 1.584E-213 | 667 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1461766 | 55.3 | 2.07E-212 | 664 |
| Agrocybe aegerita | Agrae_CAA7271705 | 56.2 | 2.763E-209 | 655 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB21378 | 53.7 | 7.778E-198 | 622 |
| Lentinula edodes NBRC 111202 | Lenedo1_1072271 | 53.2 | 4.894E-192 | 605 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_190490 | 50.4 | 2.895E-185 | 585 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_111268 | 50.4 | 6.677E-185 | 584 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_8233 | 50.5 | 1.01E-182 | 578 |
| Auricularia subglabra | Aurde3_1_1275343 | 41.4 | 5.279E-124 | 408 |
| Lentinula edodes B17 | Lened_B_1_1_1293 | 57.5 | 1.03E-100 | 339 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_492628.T0 |
| Description | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 50 | 281 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 20 | 0.9736 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR017853 | Glycoside hydrolase superfamily |
| IPR001547 | Glycoside hydrolase, family 5 |
| IPR045053 | Mannan endo-1,4-beta-mannosidase-like |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| K19355 |
EggNOG
| COG category | Description |
|---|---|
| G | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH5 | GH5_30 |
Transcription factor
| Group |
|---|
| No records |
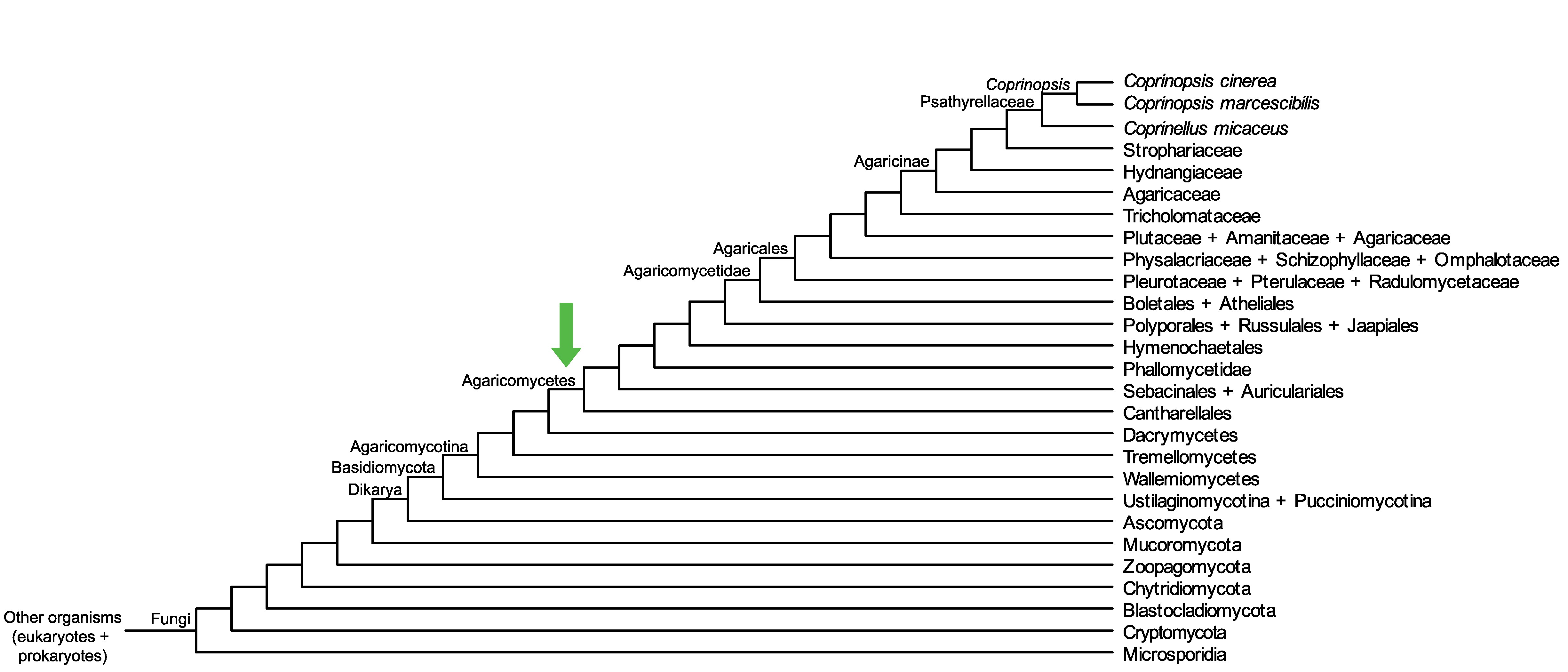
Conservation of CopciAB_492628 across fungi.
Arrow shows the origin of gene family containing CopciAB_492628.
Protein
| Sequence id | CopciAB_492628.T0 |
|---|---|
| Sequence |
>CopciAB_492628.T0 MFVAALWPVALGALAVLATAKPTRTSPLAKRQAPSDIRFVTTDNGRFVVNGAPINFVGTNAYWLHTLNSEQDIDY TLGNISAAGIKIVRTWAFNEVTSVPETGTWFQLIKDDGTVEINEGPNGLQKLDAVVRLAEKHNIYLLLALTNNWS PDPLFDDITIGAGPVRRSDITPPANGSLPRNFLSNDYGGMDTYVRQFGLDNHDEFYTNPKVINAFKNFTATIAKR YTNSPAVFGWELANDARCSSTVGATTCNPKVITKWHSNIAQHIKEVDPNHLVASGTGGQFCVDCPKLFPRVVPPP PAVSPVPGLRRRGRTGFLTKSRIIQDRLAARKRNMKRNKTKGGVKIRGRWTSSVTRRQEDLQTLGPAFDGSSGVD SEDILNIPEISFSSFQLFPDQNEYGQPDPDLSDFENTMQRGVEWIQYHAESALAFGKPATLNGFGLVTQEHVPNY VPFNSSEAPYASDTETPTEEQPYGVTNEQRDEAFSTWLNVGFRSGLSSMIHYQWGQTGLTVQPGTTVSQSVPGTG LVENDGITGVSPNDGYSSNGVGRDSVIEVLQESVQEYASDSTLR |
| Length | 569 |
Coding
| Sequence id | CopciAB_492628.T0 |
|---|---|
| Sequence |
>CopciAB_492628.T0 ATGTTCGTCGCTGCCCTTTGGCCCGTTGCTCTCGGTGCCCTCGCAGTGCTAGCGACTGCTAAGCCGACGCGAACA TCCCCTCTTGCGAAACGCCAAGCACCATCCGACATTCGGTTCGTTACGACAGATAATGGAAGATTCGTGGTTAAT GGGGCCCCAATCAACTTTGTTGGGACCAATGCCTACTGGCTTCATACCCTGAACTCTGAGCAGGATATTGACTAC ACATTGGGCAACATCTCTGCTGCAGGTATTAAGATAGTCCGCACTTGGGCGTTTAATGAAGTCACGTCTGTCCCT GAGACCGGGACATGGTTCCAGTTAATTAAAGACGACGGGACCGTTGAGATCAACGAGGGACCCAATGGCCTCCAA AAGCTGGACGCAGTCGTGAGGCTGGCTGAAAAGCATAACATCTACTTGCTTTTGGCCCTCACGAACAATTGGAGC CCCGACCCGCTCTTCGACGACATCACGATTGGTGCAGGCCCTGTTCGCCGCTCGGATATCACCCCACCAGCGAAC GGAAGTCTACCGAGAAACTTCCTCAGCAACGACTATGGTGGAATGGATACCTATGTACGACAGTTCGGATTGGAT AATCACGACGAATTCTATACGAACCCCAAAGTTATCAACGCCTTCAAGAATTTTACTGCTACGATTGCTAAACGA TACACCAATAGCCCGGCTGTCTTCGGCTGGGAGCTGGCGAACGACGCGCGCTGCAGCTCAACCGTTGGAGCTACC ACTTGCAATCCAAAGGTTATTACCAAATGGCATTCGAATATCGCCCAACATATCAAGGAAGTAGACCCTAACCAT CTGGTTGCTTCTGGAACCGGCGGACAATTCTGTGTTGACTGCCCAAAGCTTTTCCCTCGCGTCGTCCCTCCGCCT CCAGCCGTGTCCCCAGTACCTGGCCTACGCCGTCGTGGGCGCACAGGTTTCCTTACCAAATCTAGGATTATTCAA GACCGTCTCGCCGCCCGCAAGCGCAACATGAAACGCAACAAGACCAAAGGAGGTGTCAAAATCCGAGGCCGATGG ACTTCTAGTGTTACGAGGCGACAGGAAGATCTTCAAACTCTTGGGCCCGCCTTTGACGGCTCCTCCGGCGTGGAC AGTGAAGACATTTTGAACATTCCCGAAATCAGCTTCTCCTCTTTCCAACTATTCCCCGACCAGAATGAATACGGC CAACCCGACCCGGATTTGTCTGATTTCGAGAACACCATGCAGCGTGGTGTAGAATGGATCCAATATCACGCCGAA TCTGCCCTTGCTTTCGGCAAGCCTGCTACCCTAAATGGATTTGGTCTGGTCACCCAAGAGCATGTTCCTAACTAT GTCCCGTTCAACAGCTCGGAAGCTCCTTATGCTTCGGACACCGAAACACCCACAGAGGAGCAACCGTATGGAGTC ACCAACGAACAGCGTGATGAAGCCTTCTCCACCTGGTTGAATGTTGGTTTCAGGAGCGGACTTTCCAGTATGATT CACTATCAATGGGGTCAAACCGGATTGACTGTGCAGCCTGGAACCACGGTATCACAGTCTGTCCCTGGAACTGGT CTTGTCGAGAACGATGGTATCACTGGCGTCTCTCCTAACGACGGTTACTCTTCCAATGGTGTTGGACGAGATTCT GTCATTGAGGTTCTTCAAGAGAGTGTTCAAGAATACGCATCGGATTCCACCCTTCGATGA |
| Length | 1710 |
Transcript
| Sequence id | CopciAB_492628.T0 |
|---|---|
| Sequence |
>CopciAB_492628.T0 GAGCCCCCTTGCGCCCTTCCCGCAGGTTGCACGCCTAGGCTTCACAATGTTCGTCGCTGCCCTTTGGCCCGTTGC TCTCGGTGCCCTCGCAGTGCTAGCGACTGCTAAGCCGACGCGAACATCCCCTCTTGCGAAACGCCAAGCACCATC CGACATTCGGTTCGTTACGACAGATAATGGAAGATTCGTGGTTAATGGGGCCCCAATCAACTTTGTTGGGACCAA TGCCTACTGGCTTCATACCCTGAACTCTGAGCAGGATATTGACTACACATTGGGCAACATCTCTGCTGCAGGTAT TAAGATAGTCCGCACTTGGGCGTTTAATGAAGTCACGTCTGTCCCTGAGACCGGGACATGGTTCCAGTTAATTAA AGACGACGGGACCGTTGAGATCAACGAGGGACCCAATGGCCTCCAAAAGCTGGACGCAGTCGTGAGGCTGGCTGA AAAGCATAACATCTACTTGCTTTTGGCCCTCACGAACAATTGGAGCCCCGACCCGCTCTTCGACGACATCACGAT TGGTGCAGGCCCTGTTCGCCGCTCGGATATCACCCCACCAGCGAACGGAAGTCTACCGAGAAACTTCCTCAGCAA CGACTATGGTGGAATGGATACCTATGTACGACAGTTCGGATTGGATAATCACGACGAATTCTATACGAACCCCAA AGTTATCAACGCCTTCAAGAATTTTACTGCTACGATTGCTAAACGATACACCAATAGCCCGGCTGTCTTCGGCTG GGAGCTGGCGAACGACGCGCGCTGCAGCTCAACCGTTGGAGCTACCACTTGCAATCCAAAGGTTATTACCAAATG GCATTCGAATATCGCCCAACATATCAAGGAAGTAGACCCTAACCATCTGGTTGCTTCTGGAACCGGCGGACAATT CTGTGTTGACTGCCCAAAGCTTTTCCCTCGCGTCGTCCCTCCGCCTCCAGCCGTGTCCCCAGTACCTGGCCTACG CCGTCGTGGGCGCACAGGTTTCCTTACCAAATCTAGGATTATTCAAGACCGTCTCGCCGCCCGCAAGCGCAACAT GAAACGCAACAAGACCAAAGGAGGTGTCAAAATCCGAGGCCGATGGACTTCTAGTGTTACGAGGCGACAGGAAGA TCTTCAAACTCTTGGGCCCGCCTTTGACGGCTCCTCCGGCGTGGACAGTGAAGACATTTTGAACATTCCCGAAAT CAGCTTCTCCTCTTTCCAACTATTCCCCGACCAGAATGAATACGGCCAACCCGACCCGGATTTGTCTGATTTCGA GAACACCATGCAGCGTGGTGTAGAATGGATCCAATATCACGCCGAATCTGCCCTTGCTTTCGGCAAGCCTGCTAC CCTAAATGGATTTGGTCTGGTCACCCAAGAGCATGTTCCTAACTATGTCCCGTTCAACAGCTCGGAAGCTCCTTA TGCTTCGGACACCGAAACACCCACAGAGGAGCAACCGTATGGAGTCACCAACGAACAGCGTGATGAAGCCTTCTC CACCTGGTTGAATGTTGGTTTCAGGAGCGGACTTTCCAGTATGATTCACTATCAATGGGGTCAAACCGGATTGAC TGTGCAGCCTGGAACCACGGTATCACAGTCTGTCCCTGGAACTGGTCTTGTCGAGAACGATGGTATCACTGGCGT CTCTCCTAACGACGGTTACTCTTCCAATGGTGTTGGACGAGATTCTGTCATTGAGGTTCTTCAAGAGAGTGTTCA AGAATACGCATCGGATTCCACCCTTCGATGACGATAGCGCTGAAATTGCCTATTCTACTACGTCCCTGATCAGAT GACTCCGATGACGGTTTGCGCAACCTCGATCCTGCCCTGTCAGTTGCTTCTTGGACATTTCAATAATGCTTTGAA ATTTGGACACTTCACTTTGGTCGATCACGTTTGCTTTGCTTCTAATGTACAGTTAACCCCTAGCTTTCACGGATT TTATATACTATAATTCTACATCTC |
| Length | 1974 |
Gene
| Sequence id | CopciAB_492628.T0 |
|---|---|
| Sequence |
>CopciAB_492628.T0 GAGCCCCCTTGCGCCCTTCCCGCAGGTTGCACGCCTAGGCTTCACAATGTTCGTCGCTGCCCTTTGGCCCGTTGC TCTCGGTGCCCTCGCAGTGCTAGCGACTGCTAAGCCGACGCGAACATCCCCTCTTGCGAAACGCCAAGCACCATC CGACATTCGGTTCGTTACGACAGATAATGGAAGATTCGTGGTTAATGGGGCGTAAGTCGCTGCCAACTTCTCACC GGCAGATGCTCTTCTCACGAATTGTTATAGCCCAATCAACTTTGTTGGGACCAATGCCTACTGGCTTCATACCCT GAACTCTGAGCAGGATATTGACTACACATTGGGCAACATCTCTGCTGCAGGTATTAAGATAGTCCGCACTTGGGC GTTTAATGGTCCGTTCCCCCTCCCTCCCACAGGCTTATCATAAGCTAATCCAATCTCCGCATTCATTAGAAGTCA CGTCTGTCCCTGAGACCGGGACATGGTTCCAGTTAATTAAAGACGACGGGACCGTTGAGATCAACGAGGGACCCA ATGGCCTCCAAAAGCTGGACGCAGTCGTGAGGCTGGCTGAAAAGCATAACATCTACTTGCTTTTGGCCCTCACGA ACAATTGGAGCCCCGACCCGCTCTTCGACGACATCACGATTGGTGCAGGCCCTGTTCGCCGCTCGGATATCACCC CACCAGCGAACGGAAGTCTACCGAGAAACTTCCTCAGCAACGACTATGGTGCCCCAGTTCCCAACTCAAGTGGAA ACTGTATACTGACCAAATGAATGACAGGTGGAATGGATACCTATGTACGACAGTTCGGATTGGATAATCACGACG AATTCTATACGAACCCCAAAGTTATCAACGCCTTCAAGAATTTTACTGCTACGATTGCTAAACGATACACCAATA GCCCGGCTGTCTTCGGCTGGTGAGCCCTCCTTCCTCAATCAATCCAAACAATTTGGAACTGATCTTGGTTCAGGG AGCTGGCGAACGACGCGCGCTGCAGCTCAACCGTTGGAGCTACCACTTGCAATCCAAAGGTTATTACCAAATGGC ATTCGAATATCGCCCAACATATCAAGGAAGTAGACCCTAACCATCTGGTTGCTTCTGGGTAAGGCCCACCCCTGT TTGTGCGACTTGTTCACTCATCTGTGTTCTAGAACCGGCGGACAATTCTGTGTTGACTGCCCAAAGCTTTTCCCT CGCGTCGTCCCTCCGCCTCCAGCCGTGTCCCCAGTACCTGGCCTACGCCGTCGTGGGCGCACAGGTTTCCTTACC AAATCTAGGATTATTCAAGACCGTCTCGCCGCCCGCAAGCGCAACATGAAACGCAACAAGACCAAAGGAGGTGTC AAAATCCGAGGCCGATGGACTTCTAGTGGTATGTCATCCTTTGTTGCAACAATATCCGTCCCCCCAATGTGACAT CCCGTCCAGTTACGAGGCGACAGGAAGATCTTCAAACTCTTGGGCCCGCCTTTGACGGCTCCTCCGGCGTGGACA GTGAAGACATTTTGAACATTCCCGAAATCAGCTTCTCCTCTTTCCAACTATTCCCCGACCAGAATGAATACGGCC AACCCGACCCGGATTTGTCTGATTTCGAGAACACCATGCAGCGTGGTGTAGAATGGATCCAATATCACGCCGAAT CTGCCCTTGCGTAGGTTTTATCCGTTGTTTGAGCATCTAGCTTTGCTTATGCGATCTCTTTTCCAGTTTCGGCAA GCCTGCTACCCTAAATGGATTTGGTCTGGTCACCCAAGAGCATGTTCCTAACTATGTCCCGTTCAACAGCTCGGA AGCTCCTTATGCTTCGGACAGTGAGCGATTCATTGTCCCACATCATATCGTCACACTAACGTTTCTGACAGCCGA AACACCCACAGAGGAGCAACCGTATGGAGTCACCAACGAACAGCGTGATGAAGCCTTCTCCACCTGGTTGAATGT TGGTTTCAGGAGCGGACTTTCCAGTATGATTCACTATCAATGGGGTCAAACCGGATTGACTGTGCAGCCTGGAAC CACGGTATCACAGTCTGTCCCTGGAACTGGTCTTGTCGAGAACGATGGTATCACTGGCGTCTCTCCTAACGACGG TTACTCTTCCAATGGTGTTGGGTGAGTATCTGGTCTTCGCATGCAGCCTGCTCGACAAACTGACACGGACATCTA GACGAGATTCTGTCATTGAGGTTCTTCAAGAGAGTGTTCAAGAATACGCATCGGATTCCACCCTTCGATGACGAT AGCGCTGAAATTGCCTATTCTACTACGTCCCTGATCAGATGACTCCGATGACGGTTTGCGCAACCTCGATCCTGC CCTGTCAGTTGCTTCTTGGACATTTCAATAATGCTTTGAAATTTGGACACTTCACTTTGGTCGATCACGTTTGCT TTGCTTCTAATGTACAGTTAACCCCTAGCTTTCACGGATTTTATATACTATAATTCTACATCTC |
| Length | 2464 |