CopciAB_493627
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_493627 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 493627 |
| Uniprot id | Functional description | Homeodomain | |
| Location | scaffold_4:1875047..1876866 | Strand | + |
| Gene length (nt) | 1820 | Transcript length (nt) | 1582 |
| CDS length (nt) | 1161 | Protein length (aa) | 386 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7261664 | 52.7 | 4.807E-111 | 361 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB25181 | 47.4 | 4.22E-94 | 312 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_192433 | 49.8 | 1.624E-90 | 301 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_112464 | 49.8 | 1.692E-90 | 301 |
| Pleurotus ostreatus PC9 | PleosPC9_1_84519 | 43.5 | 1.096E-75 | 258 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1112690 | 43.3 | 2.465E-75 | 257 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1438647 | 43.1 | 6.387E-75 | 256 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_16333 | 43 | 7.646E-72 | 247 |
| Lentinula edodes NBRC 111202 | Lenedo1_1171205 | 41 | 5.694E-64 | 224 |
| Flammulina velutipes | Flave_chr07AA00573 | 38.5 | 1.83E-54 | 196 |
| Schizophyllum commune H4-8 | Schco3_2632356 | 37.9 | 3.445E-41 | 157 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_127053 | 33 | 6.207E-39 | 150 |
| Grifola frondosa | Grifr_OBZ68006 | 33.4 | 6.01E-31 | 126 |
| Lentinula edodes B17 | Lened_B_1_1_2729 | 80.5 | 2.159E-27 | 115 |
| Auricularia subglabra | Aurde3_1_1274371 | 30.2 | 1.261E-19 | 92 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_493627.T0 |
| Description | Homeodomain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd00086 | homeodomain | IPR001356 | 243 | 301 |
| Pfam | PF00046 | Homeodomain | IPR001356 | 243 | 299 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR001356 | Homeobox domain |
| IPR017970 | Homeobox, conserved site |
| IPR009057 | Homeobox-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0003677 | DNA binding | MF |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | MF |
| GO:0006355 | regulation of transcription, DNA-templated | BP |
KEGG
| KEGG Orthology |
|---|
| K09323 |
| K12411 |
EggNOG
| COG category | Description |
|---|---|
| K | Homeodomain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| helix-turn-helix, homeobox |
| helix-turn-helix |
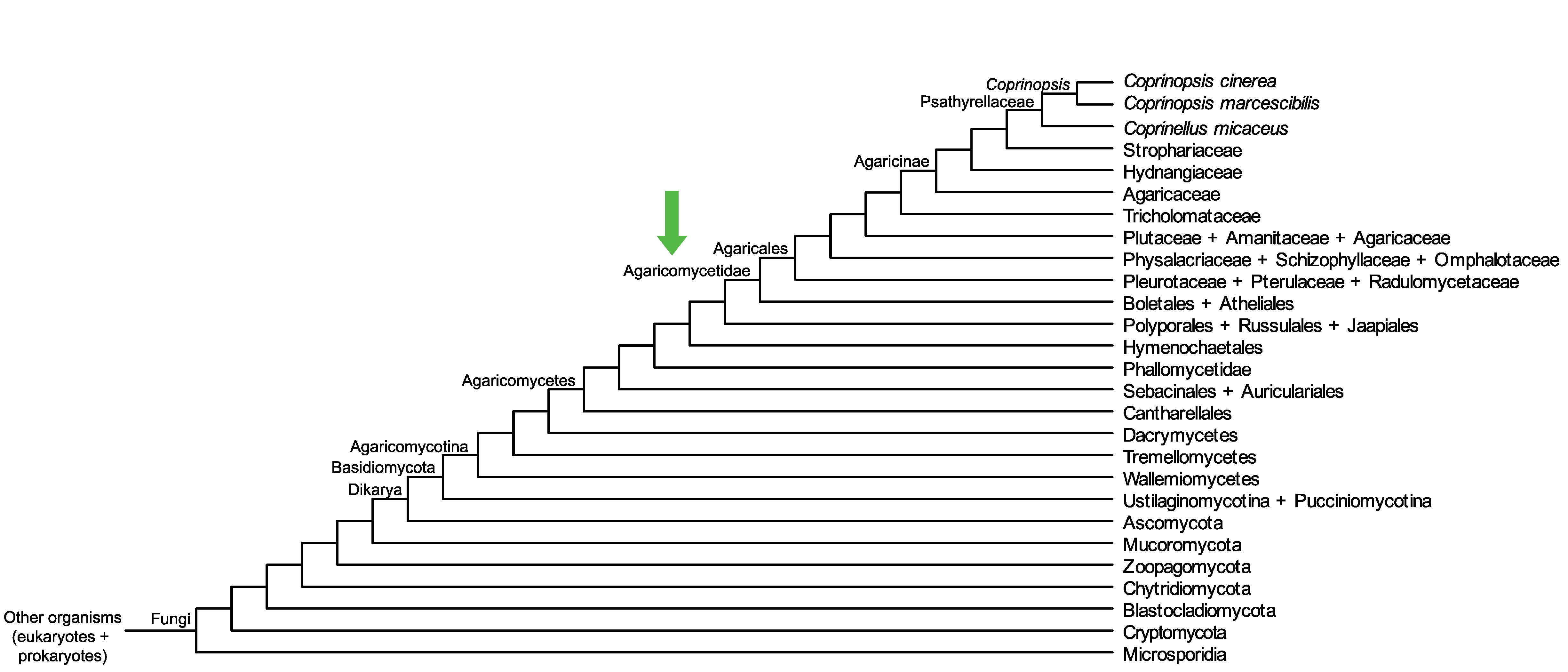
Conservation of CopciAB_493627 across fungi.
Arrow shows the origin of gene family containing CopciAB_493627.
Protein
| Sequence id | CopciAB_493627.T0 |
|---|---|
| Sequence |
>CopciAB_493627.T0 MSASANKDRYYPGTIGKSRRSSPSDTQGNPFSFQTGQGPYPNAYTAPRSSPGRGYDINPQAFYPQWPTGNSPPLT PAFPNYDTTSGHLHERHSPQPTYGYTTRTTSPPPMNPTDPRRLPPLTTSSSPGGDRWQQSANYNHGMPAVHGYPS GNSIRSPTASYPSQYVTYPSGNQGTYTYMSTYDPLAMTPQGQQGLFDDIDSVGLQQPRSTSPYGRTHAQTQLSST NFTPPPVSPISPEEPTIKKKRKRADAAQLKVLNETYARTAFPSTEERQALAKRLDMSARSVQIWFQNKRQSMRQT NRQSSTVSSSGHQQQYALQSSHNERLLDDGTHGSSYGGSITLSETPYMTGSSVQDAMTRSHTSLSSSAHRRVRGP EEDAQRWPRGY |
| Length | 386 |
Coding
| Sequence id | CopciAB_493627.T0 |
|---|---|
| Sequence |
>CopciAB_493627.T0 ATGTCGGCGTCAGCAAACAAGGATCGGTACTATCCTGGTACTATAGGGAAGTCTCGTAGAAGTTCCCCTAGTGAT ACACAAGGCAATCCGTTTTCTTTCCAGACGGGACAGGGGCCCTATCCTAATGCATATACCGCGCCTCGCTCATCT CCAGGTAGAGGGTACGACATCAATCCTCAAGCGTTTTACCCCCAATGGCCGACTGGGAACTCCCCGCCACTCACC CCCGCATTCCCCAACTATGACACGACGAGCGGCCATCTCCACGAACGACACTCTCCGCAACCTACTTACGGTTAC ACTACTCGGACAACCTCGCCTCCTCCGATGAACCCTACAGATCCTCGCAGGCTCCCTCCCCTCACGACGTCGTCG AGCCCTGGTGGCGATCGATGGCAACAGTCGGCCAACTACAACCATGGTATGCCCGCTGTCCATGGCTACCCCTCC GGCAATAGCATTCGCTCGCCGACTGCGAGCTACCCTTCGCAGTATGTTACCTATCCCAGCGGTAACCAAGGTACT TACACCTATATGTCAACATACGATCCGCTCGCGATGACCCCCCAAGGCCAACAAGGCCTCTTTGACGATATCGAC TCTGTCGGACTCCAACAACCTCGATCAACCTCGCCTTATGGGCGTACCCATGCCCAAACGCAGTTGTCTTCTACC AATTTCACCCCTCCACCGGTATCGCCCATTTCCCCGGAGGAGCCCACCATCAAGAAAAAGCGCAAGCGTGCCGAC GCTGCTCAGCTCAAGGTCCTCAATGAGACCTATGCCCGCACCGCTTTCCCATCCACGGAAGAACGGCAGGCCCTT GCAAAGCGTTTGGACATGTCGGCCAGAAGTGTTCAGATTTGGTTCCAGAACAAGCGGCAGTCGATGAGGCAGACG AACCGCCAATCGTCTACCGTTTCGTCTTCTGGCCATCAACAGCAGTACGCGCTGCAGAGCTCTCACAACGAGAGG TTGTTGGACGATGGGACCCATGGATCTTCTTACGGCGGCTCTATCACTTTGTCGGAGACCCCTTACATGACAGGA AGCTCTGTGCAAGACGCGATGACACGTTCACATACCTCCCTTTCATCTTCTGCGCATCGCCGAGTGCGAGGCCCT GAAGAAGACGCTCAACGGTGGCCTCGCGGTTACTGA |
| Length | 1161 |
Transcript
| Sequence id | CopciAB_493627.T0 |
|---|---|
| Sequence |
>CopciAB_493627.T0 ATTAATATCACTTCCCCTCTTCCCCGCTTCTTATTACTACTTTTCTCTTTCTCCACTAATATGTCGGCGTCAGCA AACAAGGATCGGTACTATCCTGGTACTATAGGGAAGTCTCGTAGAAGTTCCCCTAGTGATACACAAGGCAATCCG TTTTCTTTCCAGACGGGACAGGGGCCCTATCCTAATGCATATACCGCGCCTCGCTCATCTCCAGGTAGAGGGTAC GACATCAATCCTCAAGCGTTTTACCCCCAATGGCCGACTGGGAACTCCCCGCCACTCACCCCCGCATTCCCCAAC TATGACACGACGAGCGGCCATCTCCACGAACGACACTCTCCGCAACCTACTTACGGTTACACTACTCGGACAACC TCGCCTCCTCCGATGAACCCTACAGATCCTCGCAGGCTCCCTCCCCTCACGACGTCGTCGAGCCCTGGTGGCGAT CGATGGCAACAGTCGGCCAACTACAACCATGGTATGCCCGCTGTCCATGGCTACCCCTCCGGCAATAGCATTCGC TCGCCGACTGCGAGCTACCCTTCGCAGTATGTTACCTATCCCAGCGGTAACCAAGGTACTTACACCTATATGTCA ACATACGATCCGCTCGCGATGACCCCCCAAGGCCAACAAGGCCTCTTTGACGATATCGACTCTGTCGGACTCCAA CAACCTCGATCAACCTCGCCTTATGGGCGTACCCATGCCCAAACGCAGTTGTCTTCTACCAATTTCACCCCTCCA CCGGTATCGCCCATTTCCCCGGAGGAGCCCACCATCAAGAAAAAGCGCAAGCGTGCCGACGCTGCTCAGCTCAAG GTCCTCAATGAGACCTATGCCCGCACCGCTTTCCCATCCACGGAAGAACGGCAGGCCCTTGCAAAGCGTTTGGAC ATGTCGGCCAGAAGTGTTCAGATTTGGTTCCAGAACAAGCGGCAGTCGATGAGGCAGACGAACCGCCAATCGTCT ACCGTTTCGTCTTCTGGCCATCAACAGCAGTACGCGCTGCAGAGCTCTCACAACGAGAGGTTGTTGGACGATGGG ACCCATGGATCTTCTTACGGCGGCTCTATCACTTTGTCGGAGACCCCTTACATGACAGGAAGCTCTGTGCAAGAC GCGATGACACGTTCACATACCTCCCTTTCATCTTCTGCGCATCGCCGAGTGCGAGGCCCTGAAGAAGACGCTCAA CGGTGGCCTCGCGGTTACTGAGATTCAGGATGCTTGATTCACTTTTCATTCTGGGTTCGCATAGTTACATCCGCT CACAAACTTCTTCCATCGGGCTTCTTTTTATAACCATAGAGACTTTGACTCTAATTTACAGCGTCAGTATTAATT GGTTCATTAATATCTCCTACTCCTTCTACTTATTGCTCTGTCTTGACAGGGCGCGGCAGCTTTTTATTTTCTTAT TTGGCTTCATTTTATTCTCTTATCTAACGACCTCGTCATGCTTCCTATACCCACCGCACTCCCATGCAGACATGC AACATTTCTTATGGCACTTTGGATCACTCCATGTATCACACTTCTTTCTGTACTTATCCAATTCTATGCCTTATG CCCACGC |
| Length | 1582 |
Gene
| Sequence id | CopciAB_493627.T0 |
|---|---|
| Sequence |
>CopciAB_493627.T0 ATTAATATCACTTCCCCTCTTCCCCGCTTCTTATTACTACTTTTCTCTTTCTCCACTAATATGTCGGCGTCAGCA AACAAGGATCGGTACTATCCTGGTACTATAGGGAAGTCTCGTAGAAGTTCCCCTAGTGATACACAAGGCAATCCG TTTTCTTTCCAGACGGGACAGGGTGGGCGAACAGTGTTGCCTTCTTTCTCTACGGCCTTTCCAAACTTACCTTTT CCAGGTCTGTCTCTTTTTGTAGTTTTAAGCCTTCCGAAAACACTCAAACCTTCATCTTTCATCGTTACAGGGCCC TATCCTAATGCATATACCGCGCCTCGCTCATCTCCAGGTAGAGGGTACGACATCAATCCTCAAGCGTTTTACCCC CAATGGCCGACTGGGAACTGTACGTTCATTAATTTCCTTTTTTCCATGGTGTTCAAATTATCTCCATTGGCGCCA ATAGCCCCGCCACTCACCCCCGCATTCCCCAACTATGACACGACGAGCGGCCATCTCCACGAACGACACTCTCCG CAACCTACTTACGGTTACACTACTCGGACAACCTCGCCTCCTCCGATGAACCCTACAGATCCTCGCAGGCTCCCT CCCCTCACGACGTCGTCGAGCCCTGGTGGCGATCGATGGCAACAGTCGGCCAACTACAACCATGGTATGCCCGCT GTCCATGGCTACCCCTCCGGCAATAGCATTCGCTCGCCGACTGCGAGCTACCCTTCGCAGTATGTTACCTATCCC AGCGGTAACCAAGGTACTTACACCTATATGTCAACATACGATCCGCTCGCGATGACCCCCCAAGGCCAACAAGGC CTCTTTGACGATATCGACTCTGTCGGACTCCAACAACCTCGATCAACCTCGCCTTATGGGCGTACCCATGCCCAA ACGCAGTTGTCTTCTACCAATTTCACCCCTCCACCGGTATCGCCCATTTCCCCGGAGGAGCCCACCATCAAGAAA AAGCGCAAGCGTGCCGACGCTGCTCAGCTCAAGGTCCTCAATGAGACCTATGCCCGCACCGCTTTCCCATCCACG GAAGAACGGCAGGCCCTTGCAAAGCGTTTGGACATGTCGGCCAGAAGTGTTCAGATTTGGTACGTCATCATGGTG CCTTATATATTTTTGCCATCCGTATTAACAAGGATCCAGGTTCCAGAACAAGCGGCAGTCGATGAGGCAGACGAA CCGCCAATCGTCTACCGTTTCGTCTTCTGGCCATCAACAGCAGTACGCGCTGCAGAGCTCTCACAACGAGAGGTT GTTGGACGATGGGACCCATGGATCTTCTTACGGCGGCTCTATCACTTTGTCGGAGACCCCTTACATGACAGGAAG CTCTGTGCAAGACGCGATGACACGTTCACATACCTCCCTTTCATCTTCTGCGCATCGCCGAGTGCGAGGCCCTGA AGAAGACGCTCAACGGTGGCCTCGCGGTTACTGAGATTCAGGATGCTTGATTCACTTTTCATTCTGGGTTCGCAT AGTTACATCCGCTCACAAACTTCTTCCATCGGGCTTCTTTTTATAACCATAGAGACTTTGACTCTAATTTACAGC GTCAGTATTAATTGGTTCATTAATATCTCCTACTCCTTCTACTTATTGCTCTGTCTTGACAGGGCGCGGCAGCTT TTTATTTTCTTATTTGGCTTCATTTTATTCTCTTATCTAACGACCTCGTCATGCTTCCTATACCCACCGCACTCC CATGCAGACATGCAACATTTCTTATGGCACTTTGGATCACTCCATGTATCACACTTCTTTCTGTACTTATCCAAT TCTATGCCTTATGCCCACGC |
| Length | 1820 |