CopciAB_494762
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_494762 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 494762 |
| Uniprot id | Functional description | Polysaccharide lyase family 8, N terminal alpha-helical domain | |
| Location | scaffold_1:2629767..2632914 | Strand | - |
| Gene length (nt) | 3148 | Transcript length (nt) | 2533 |
| CDS length (nt) | 2442 | Protein length (aa) | 813 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB29606 | 57 | 3.272E-293 | 911 |
| Agrocybe aegerita | Agrae_CAA7266471 | 53.5 | 6.629E-273 | 852 |
| Grifola frondosa | Grifr_OBZ79298 | 50.8 | 5.764E-253 | 794 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1476771 | 51 | 3.097E-252 | 792 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_214326 | 51.4 | 2.656E-250 | 786 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_31645 | 51.3 | 2.99E-249 | 783 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1111478 | 50 | 1.624E-248 | 781 |
| Pleurotus ostreatus PC9 | PleosPC9_1_53101 | 49.8 | 1.717E-247 | 778 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_124358 | 49 | 6.094E-242 | 762 |
| Flammulina velutipes | Flave_chr08AA00957 | 48.5 | 5.908E-232 | 733 |
| Schizophyllum commune H4-8 | Schco3_2628008 | 45.1 | 6.907E-211 | 672 |
| Auricularia subglabra | Aurde3_1_1244861 | 43.8 | 3.316E-204 | 653 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_494762.T0 |
| Description | Polysaccharide lyase family 8, N terminal alpha-helical domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF08124 | Polysaccharide lyase family 8, N terminal alpha-helical domain | IPR012970 | 203 | 354 |
| Pfam | PF02278 | Polysaccharide lyase family 8, super-sandwich domain | IPR003159 | 438 | 684 |
| Pfam | PF02884 | Polysaccharide lyase family 8, C-terminal beta-sandwich domain | IPR004103 | 700 | 771 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 27 | 0.5009 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| 1 | 7 | 26 | 19 |
InterPro
| Accession | Description |
|---|---|
| IPR003159 | Polysaccharide lyase family 8, central domain |
| IPR008929 | Chondroitin AC/alginate lyase |
| IPR012970 | Polysaccharide lyase 8, N-terminal alpha-helical |
| IPR004103 | Polysaccharide lyase family 8, C-terminal |
| IPR014718 | Glycoside hydrolase-type carbohydrate-binding |
| IPR011071 | Polysaccharide lyase family 8-like, C-terminal |
| IPR011013 | Galactose mutarotase-like domain superfamily |
| IPR038970 | Polysaccharide lyase 8 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005576 | extracellular region | CC |
| GO:0016829 | lyase activity | MF |
| GO:0030246 | carbohydrate binding | MF |
| GO:0005975 | carbohydrate metabolic process | BP |
| GO:0003824 | catalytic activity | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| G | Polysaccharide lyase family 8, N terminal alpha-helical domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| PL | PL8 | PL8_4 |
Transcription factor
| Group |
|---|
| No records |
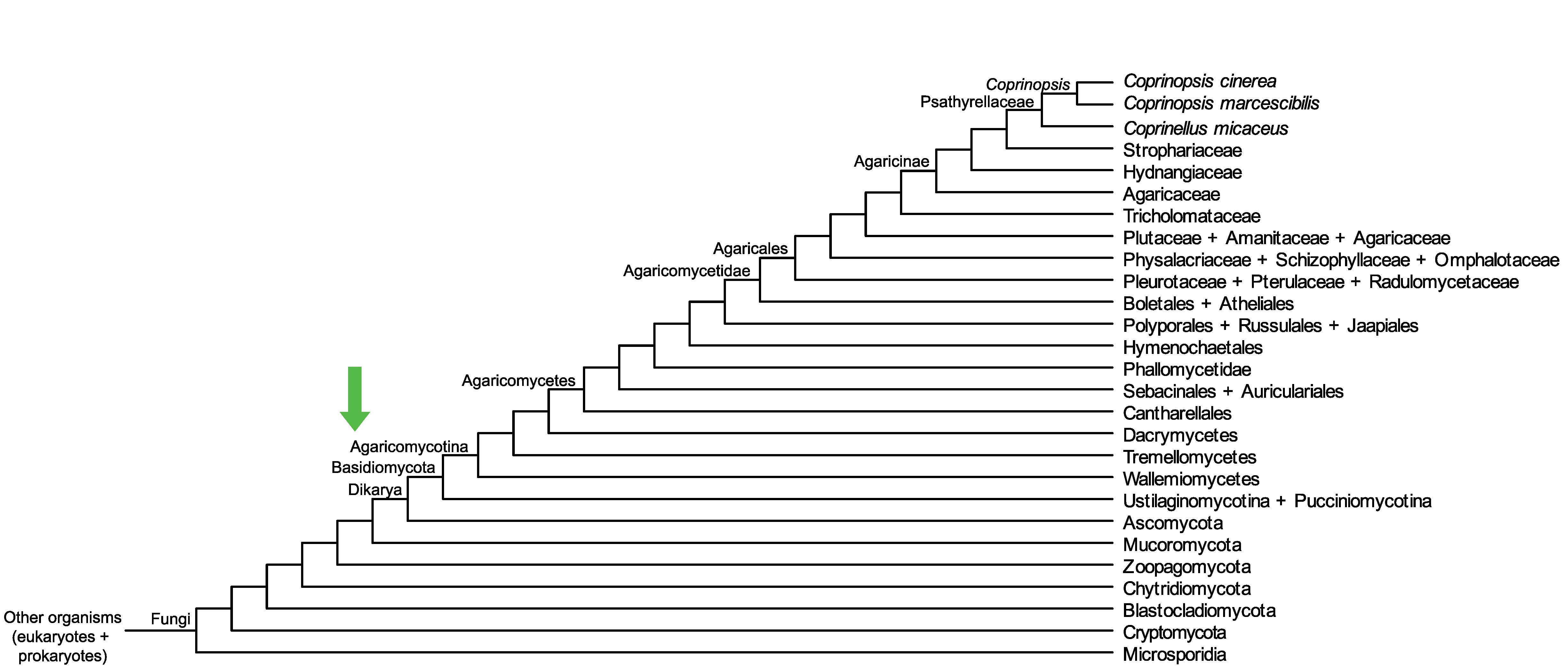
Conservation of CopciAB_494762 across fungi.
Arrow shows the origin of gene family containing CopciAB_494762.
Protein
| Sequence id | CopciAB_494762.T0 |
|---|---|
| Sequence |
>CopciAB_494762.T0 MMRFGSAVALSGCLLSMLLIFSSLLLPSEFPSRSLSGWLAAVKGRGAFDYLLRHGSGPSPMQYRSASKITRQVSE DLSVLHQRRVSLIVGQLRNPSNISHWLDSLDNLTGKWPDTEVDYTTGCGARRANWPAQAHWQRILVMSGAWHGGY RGAEQWVKDERLRTTIGRAMEYWFSRDITNVDCLDWGGTPRCPCDNVDDSLWNTNWFSNIILIPRLAGQSCLLLS DSLTLREREVCSNITRRSFGTFGIKLNQVGYLTGANTLDVARVGIDCGLLNSNISMIADAYRRIHDELVIQNVTK VDGIRADGSFAQHAGVLYNGNYGKDYSNAVLDVEVIAAGTSFSADTTAQTAFETLFDGHRWMIFSNTLSQVLHFD FSVLGRFITFPVIDDQATGSIQINLTKTWELGRLWSSPSLTTFGESLMQNFSSANAGGLTGTRMFFANDYMVHRG PNFFSSLKMYSSRTLNTECINSQNPLGFHLSDGTLYTYLRGNEYTDISAAWDWDLIPGTTVDYRNTLLHCSGATV TGRESFVGGLSDGKTGIAAMRYTNPTTRSLRWQKTWFFLNDGAQHVVINILASSSGAPVYSVLDQKRRNGLTVVD RREVLNSSTITGPQRLWHDNVGYAFRSLTNATTPLHVRMGERTGSWSAIGTSTQPPVSVDMWAGLLEHIDLSQPL EYTILPGTSYPDFLQRTELPRVETIANNDNVTAIYDQRTGIAMVVFWNNGESSIAIDTRTPIAQMTITSTANVIV MFSTKNGTVLVSDPTQMLDTVVLKFTIGRGKRPLYWKGQAEKSLVFQLPGGGRRGDSVSGTIR |
| Length | 813 |
Coding
| Sequence id | CopciAB_494762.T0 |
|---|---|
| Sequence |
>CopciAB_494762.T0 ATGATGCGTTTCGGTTCTGCGGTGGCACTGAGTGGGTGCTTGCTTTCAATGCTCCTCATTTTCTCCTCCCTCCTC CTGCCTTCCGAGTTTCCCTCGCGATCGCTCTCGGGCTGGCTTGCGGCTGTGAAAGGTCGGGGGGCCTTCGACTAC CTACTGCGCCATGGTTCAGGTCCCTCCCCCATGCAATATCGGAGCGCTTCCAAGATTACTAGACAGGTCTCGGAG GATCTGTCGGTTCTTCACCAACGTCGGGTTTCCTTGATAGTCGGTCAGCTCAGAAATCCTTCGAACATATCCCAT TGGCTTGACTCGCTGGACAATTTGACAGGCAAATGGCCTGATACAGAAGTTGATTACACGACCGGCTGCGGGGCG CGAAGGGCTAACTGGCCTGCTCAGGCGCATTGGCAGAGGATCCTGGTCATGTCCGGGGCTTGGCACGGAGGATAC AGAGGAGCAGAACAATGGGTCAAGGACGAAAGGCTGAGAACTACCATTGGACGAGCCATGGAATATTGGTTCTCC CGTGATATCACGAATGTTGACTGTCTCGATTGGGGTGGAACCCCCCGCTGCCCGTGTGACAACGTCGATGATTCT CTATGGAATACCAACTGGTTCTCAAACATTATCCTCATTCCCAGACTCGCAGGACAATCATGTCTGCTCTTAAGC GATTCGCTGACCTTGAGGGAAAGAGAAGTTTGTTCGAACATTACTCGACGATCGTTTGGAACCTTTGGGATTAAG CTGAATCAGGTTGGATACCTGACTGGCGCGAATACACTCGATGTAGCACGAGTCGGAATTGACTGCGGGCTGCTC AATAGCAATATTTCAATGATCGCCGACGCTTATCGTCGAATCCACGACGAATTGGTCATTCAAAACGTCACAAAA GTAGACGGAATACGGGCAGACGGTTCCTTTGCGCAACATGCAGGTGTTCTTTACAATGGAAACTATGGAAAGGAC TACTCGAACGCAGTGTTGGACGTCGAGGTGATTGCGGCGGGAACATCGTTCTCGGCAGATACGACTGCTCAGACC GCATTCGAGACGTTGTTCGACGGACATAGGTGGATGATCTTCTCCAATACGCTTTCCCAGGTCTTGCACTTTGAC TTTTCCGTTCTTGGGCGATTTATCACGTTCCCTGTCATCGACGACCAGGCTACTGGGAGCATCCAAATCAACCTC ACCAAAACTTGGGAACTAGGGCGCCTATGGTCTTCCCCATCCTTGACTACATTCGGGGAGTCACTCATGCAGAAC TTCTCCAGCGCAAACGCAGGAGGATTGACGGGAACGAGAATGTTCTTTGCAAATGATTATATGGTCCACAGAGGT CCAAACTTTTTCTCGTCTCTCAAGATGTACTCCAGTCGAACGCTTAACACAGAATGCATCAACTCACAGAATCCG CTCGGGTTCCACCTCTCCGATGGAACGCTTTACACTTACCTCCGAGGAAATGAATATACCGATATCTCTGCTGCA TGGGACTGGGATCTGATTCCCGGAACAACAGTGGATTACCGCAACACGCTTTTACATTGCAGTGGGGCCACGGTC ACAGGCCGGGAGAGTTTCGTTGGCGGTTTATCCGATGGAAAGACTGGAATAGCGGCCATGCGTTATACGAACCCC ACCACTCGATCGCTGCGATGGCAAAAGACATGGTTCTTCCTCAACGACGGTGCCCAACACGTCGTGATCAACATT CTGGCGTCCAGTAGCGGGGCCCCCGTATATTCAGTTTTGGATCAAAAGCGTCGCAACGGACTCACCGTCGTAGAC AGGAGGGAAGTATTGAACTCGTCGACTATCACTGGCCCTCAACGGCTATGGCACGACAATGTCGGGTATGCCTTC AGGAGCTTGACAAATGCCACTACTCCCTTGCACGTACGCATGGGCGAGAGGACTGGGAGCTGGTCGGCGATTGGT ACGTCGACCCAACCCCCGGTGTCTGTTGACATGTGGGCAGGGTTGCTGGAACATATCGACCTGTCGCAACCTCTG GAATATACCATCCTTCCTGGAACTTCATACCCAGACTTCCTGCAGAGAACCGAATTACCCAGAGTGGAGACCATC GCCAACAACGACAACGTTACCGCTATTTACGATCAGCGAACGGGAATCGCGATGGTCGTATTTTGGAACAACGGC GAAAGTTCAATTGCAATCGATACCCGGACACCCATTGCTCAAATGACCATTACATCCACAGCCAATGTCATTGTA ATGTTTTCCACCAAAAACGGCACAGTGCTTGTCTCCGACCCGACCCAAATGCTTGACACAGTCGTACTGAAATTC ACTATCGGCCGGGGCAAACGACCTTTGTACTGGAAGGGGCAGGCGGAAAAATCGCTGGTATTTCAGTTGCCTGGT GGAGGACGGAGGGGAGACAGTGTATCTGGCACCATTCGATAA |
| Length | 2442 |
Transcript
| Sequence id | CopciAB_494762.T0 |
|---|---|
| Sequence |
>CopciAB_494762.T0 AGCCTAGCCTTTCCTACCCTGCGCCTTTCACCACTGCAATATTTGGTCACAACCCTTTCTTGACATGATGCGTTT CGGTTCTGCGGTGGCACTGAGTGGGTGCTTGCTTTCAATGCTCCTCATTTTCTCCTCCCTCCTCCTGCCTTCCGA GTTTCCCTCGCGATCGCTCTCGGGCTGGCTTGCGGCTGTGAAAGGTCGGGGGGCCTTCGACTACCTACTGCGCCA TGGTTCAGGTCCCTCCCCCATGCAATATCGGAGCGCTTCCAAGATTACTAGACAGGTCTCGGAGGATCTGTCGGT TCTTCACCAACGTCGGGTTTCCTTGATAGTCGGTCAGCTCAGAAATCCTTCGAACATATCCCATTGGCTTGACTC GCTGGACAATTTGACAGGCAAATGGCCTGATACAGAAGTTGATTACACGACCGGCTGCGGGGCGCGAAGGGCTAA CTGGCCTGCTCAGGCGCATTGGCAGAGGATCCTGGTCATGTCCGGGGCTTGGCACGGAGGATACAGAGGAGCAGA ACAATGGGTCAAGGACGAAAGGCTGAGAACTACCATTGGACGAGCCATGGAATATTGGTTCTCCCGTGATATCAC GAATGTTGACTGTCTCGATTGGGGTGGAACCCCCCGCTGCCCGTGTGACAACGTCGATGATTCTCTATGGAATAC CAACTGGTTCTCAAACATTATCCTCATTCCCAGACTCGCAGGACAATCATGTCTGCTCTTAAGCGATTCGCTGAC CTTGAGGGAAAGAGAAGTTTGTTCGAACATTACTCGACGATCGTTTGGAACCTTTGGGATTAAGCTGAATCAGGT TGGATACCTGACTGGCGCGAATACACTCGATGTAGCACGAGTCGGAATTGACTGCGGGCTGCTCAATAGCAATAT TTCAATGATCGCCGACGCTTATCGTCGAATCCACGACGAATTGGTCATTCAAAACGTCACAAAAGTAGACGGAAT ACGGGCAGACGGTTCCTTTGCGCAACATGCAGGTGTTCTTTACAATGGAAACTATGGAAAGGACTACTCGAACGC AGTGTTGGACGTCGAGGTGATTGCGGCGGGAACATCGTTCTCGGCAGATACGACTGCTCAGACCGCATTCGAGAC GTTGTTCGACGGACATAGGTGGATGATCTTCTCCAATACGCTTTCCCAGGTCTTGCACTTTGACTTTTCCGTTCT TGGGCGATTTATCACGTTCCCTGTCATCGACGACCAGGCTACTGGGAGCATCCAAATCAACCTCACCAAAACTTG GGAACTAGGGCGCCTATGGTCTTCCCCATCCTTGACTACATTCGGGGAGTCACTCATGCAGAACTTCTCCAGCGC AAACGCAGGAGGATTGACGGGAACGAGAATGTTCTTTGCAAATGATTATATGGTCCACAGAGGTCCAAACTTTTT CTCGTCTCTCAAGATGTACTCCAGTCGAACGCTTAACACAGAATGCATCAACTCACAGAATCCGCTCGGGTTCCA CCTCTCCGATGGAACGCTTTACACTTACCTCCGAGGAAATGAATATACCGATATCTCTGCTGCATGGGACTGGGA TCTGATTCCCGGAACAACAGTGGATTACCGCAACACGCTTTTACATTGCAGTGGGGCCACGGTCACAGGCCGGGA GAGTTTCGTTGGCGGTTTATCCGATGGAAAGACTGGAATAGCGGCCATGCGTTATACGAACCCCACCACTCGATC GCTGCGATGGCAAAAGACATGGTTCTTCCTCAACGACGGTGCCCAACACGTCGTGATCAACATTCTGGCGTCCAG TAGCGGGGCCCCCGTATATTCAGTTTTGGATCAAAAGCGTCGCAACGGACTCACCGTCGTAGACAGGAGGGAAGT ATTGAACTCGTCGACTATCACTGGCCCTCAACGGCTATGGCACGACAATGTCGGGTATGCCTTCAGGAGCTTGAC AAATGCCACTACTCCCTTGCACGTACGCATGGGCGAGAGGACTGGGAGCTGGTCGGCGATTGGTACGTCGACCCA ACCCCCGGTGTCTGTTGACATGTGGGCAGGGTTGCTGGAACATATCGACCTGTCGCAACCTCTGGAATATACCAT CCTTCCTGGAACTTCATACCCAGACTTCCTGCAGAGAACCGAATTACCCAGAGTGGAGACCATCGCCAACAACGA CAACGTTACCGCTATTTACGATCAGCGAACGGGAATCGCGATGGTCGTATTTTGGAACAACGGCGAAAGTTCAAT TGCAATCGATACCCGGACACCCATTGCTCAAATGACCATTACATCCACAGCCAATGTCATTGTAATGTTTTCCAC CAAAAACGGCACAGTGCTTGTCTCCGACCCGACCCAAATGCTTGACACAGTCGTACTGAAATTCACTATCGGCCG GGGCAAACGACCTTTGTACTGGAAGGGGCAGGCGGAAAAATCGCTGGTATTTCAGTTGCCTGGTGGAGGACGGAG GGGAGACAGTGTATCTGGCACCATTCGATAACTTTTCAATCCACATTGGTTGATCATG |
| Length | 2533 |
Gene
| Sequence id | CopciAB_494762.T0 |
|---|---|
| Sequence |
>CopciAB_494762.T0 AGCCTAGCCTTTCCTACCCTGCGCCTTTCACCACTGCAATATTTGGTCACAACCCTTTCTTGACATGATGCGTTT CGGTTCTGCGGTGGCACTGAGTGGGTGCTTGCTTTCAATGCTCCTCATTTTCTCCTCCCTCCTCCTGCCTTCCGA GTTTCCCTCGCGATCGCTCTCGGGCTGGCTTGCGGCTGTGAAAGGTCGGGGGGCCTTCGACTACCTACTGCGCCA TGGTTCAGGTCCCTCCCCCATGCAATATCGGAGCGCTTCCAAGATTACTAGACAGGTCTCGGAGGATCTGTCGGT TCTTCACCAACGTCGGGTTTCCTTGATAGTCGGTCAGCTCAGAAATCCTTCGAACATATCCCATTGGTGAGTCAT GACATTGCCAATGGCTCTAGCGGGTGTTCACAAATGGGAATTGGCTCGTTGTAGGCTTGACTCGCTGGACAATTT GACAGGCAAATGGCCTGATACAGAAGTTGATTACACGACCGGCTGCGGGGCGCGAAGGGCTAACTGGCCTGCTCA GGCGCATTGGCAGAGGATCCGTGCGTCATACATGCAATATGCCATTTAATCTTTGGTTCTGACATCTGGACAGTG GTCATGTCCGGGGCTTGGCACGGAGGATACAGAGGAGCAGAACAATGGGTCAAGGACGAAAGGCTGAGAACTACC ATTGGACGAGCCATGGAATATTGGTTCTCCCGTGATATCACGAATGTTGACTGTCTCGATTGGGGTGGAACCCCC CGCTGCCCGTGTGACAACGTCGATGATTCTCTATGGTAAGCCGTCTTCCCGTCATCTGCATCTAAATGTCTCTCT GAAGGGCCACTCCACAGGAATACCAACTGGTTCTCAAACGTAAGCTTAAAGTCACTCCATGTCTTGATGAAGATA ACGTACTGACCTCGCAAAACAGATTATCCTCATTCCCAGACTCGCAGGACAATCATGTCTGCTCTTAAGCGATTC GCTGACCTTGAGGGAAAGAGAAGTTTGTTCGAACATTACTCGACGATCGTTTGGAACCTTTGGGATTAAGCTGAA TCAGGTTGGATACCTGACTGGCGCGAATACACTCGATGTAGCACGAGTCGGAATTGACTGCGGGCTGCTCAATAG CAATATTTCAATGATCGCCGACGCTTATCGTCGAATCCACGACGAATTGGTCATTCAAAACGTCACAAAAGTAGA CGGAATACGGGCAGACGGTTCCTTTGGTGCGTCTATGCGTTCACTTCACGAACGTATTTCAAACATATTTCATCC AGCGCAACATGCAGGTGTTCTTTACAATGGAAACTATGGTACGTTCTATGCCACGTGGTATATTGTCTAAACCAA TTAAGGTGCGACCAGGAAAGGACTAGTAAGGGGTTGACAGTTCTGTCGACCTTGCCAGACAATGAACACTCCTCT TAGCTCGAACGCAGTGTTGGACGTCGAGGTGATTGCGGCGGGAACATCGTTCTCGGCAGATACGACTGCTCAGAC CGCATTCGAGACGTTGTTCGACGGACATAGGTGGATGATCTTCTCCAATACGCTTTCCCAGGTCTTGCACTTTGA CTTTGTGAGTTCGTTCGCATTCGCGCAAGCTGCGACTCATGAGGCCGTCCTTCCAACTAGTCCGTTCTTGGGCGA TTTATCACGTTCCCTGTCATCGACGACCAGTAAGTCCAAACCTCAATAGCCTGTCAAATATCATGTCTGAGCGTT TCTAATAGGGCTACTGGGAGCATCCAAATCAACCTCACCAAAACTTGGGAACTAGGGCGCCTATGGTCTTCCCCA TCCTTGACTACATTCGGGGAGTCACTCATGCAGAACTTCTCCAGCGCAAACGCAGGAGGATTGACGGGAACGAGA ATGTTCTTTGCAAATGATTATATGGTGGGCCACCACATTCACTAGCAGCTCGCTCAACCGGATTCGAGTCTATGA TTCACAGGTCCACAGAGGTCCAAACTTTTTCTCGTCTCTCAAGATGTACTCCAGTCGAACGCTTAACACAGAATG CATCAACTCACAGAATGTAAGTCGTATTTTAACGAACGTATGTTATCTGGGCTGATCCTTGCTCCGATCCGATTA GCCGCTCGGGTTCCACCTCTCCGATGGAACGCTTTACACTTACCTCCGAGGAAATGAATATACCGATATCTCTGC TGCATGGGACTGGGATCTGATTCCCGGAACAACAGTGGATTACCGCAACACGCTTTTACATTGCAGTGGGGCCAC GGTCACAGGCCGGGAGAGTTTCGTTGGCGGTTTATCCGATGGAAAGACTGGAATAGCGGCCATGCGTTATACGAA CCCCACCACTCGATCGCTGCGATGGCAAAAGACATGGTTCTTCCTCAACGACGGTGCCCAACACGTCGTGATCAA CATTCTGGCGTCCAGTAGCGGGGCCCCCGTATATTCAGTTTTGGATCAAAAGCGTCGCAACGGACTCACCGTCGT AGACAGGAGGGAAGTATTGAACTCGTCGACTATCACTGGCCCTCAACGGCTATGGCACGACAATGTCGGGTATGC CTTCAGGAGCTTGACAAATGCCACTACTCCCTTGCACGTACGCATGGGCGAGAGGACTGGGAGCTGGTCGGCGAT TGGTACGTCGACCCAACCCCCGGTGTCTGTTGACATGTGGGCAGGGTTGCTGGAACATATCGACCTGTCGCAACC TCTGGAATATACCATCCTTCCTGGAACTTCATACCCAGACTTCCTGCAGAGAACCGAATTACCCAGAGTGGAGAC CATCGCCAACAACGACAACGTTACCGCTATTTACGATCAGCGAACGGGAATCGCGATGGTCGTATTTTGGAACAA CGGCGAAAGTTCAATTGCAATCGATACCCGGACACCCATTGCTCAAATGACCATTACATCCACAGCCAATGTCAT TGTAATGTTTTCCACCAAAAACGGCACAGTGCTTGTCTCCGACCCGACCCAAATGCTTGACACAGTCGTACTGAA ATTCACTATCGGCCGGGGCAAACGACCTTTGTACTGGAAGGGGCAGGCGGAAAAATCGCTGGTATTTCAGTTGCC TGGTGGAGGACGGAGGGGAGACAGTGTATCTGGCACCATTCGATAACTTTTCAATCCACATTGGTTGATCATG |
| Length | 3148 |