CopciAB_497514
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_497514 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 497514 |
| Uniprot id | Functional description | Glycosyl hydrolase family 85 | |
| Location | scaffold_5:2948332..2951075 | Strand | + |
| Gene length (nt) | 2744 | Transcript length (nt) | 2532 |
| CDS length (nt) | 2364 | Protein length (aa) | 787 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7269656 | 46.5 | 2.613E-231 | 730 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB20368 | 44.6 | 8.4E-217 | 688 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_11491 | 42.3 | 5.703E-198 | 633 |
| Lentinula edodes NBRC 111202 | Lenedo1_1235929 | 42.3 | 2.795E-197 | 631 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_80764 | 41.7 | 3.881E-191 | 613 |
| Pleurotus ostreatus PC9 | PleosPC9_1_115065 | 40.4 | 4.65E-187 | 601 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1092656 | 41.4 | 2.306E-186 | 599 |
| Schizophyllum commune H4-8 | Schco3_2681989 | 40.9 | 1.693E-183 | 591 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1386094 | 42.8 | 1.831E-180 | 582 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_181728 | 39.4 | 8.994E-175 | 565 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_109029 | 39.5 | 4.538E-174 | 563 |
| Lentinula edodes B17 | Lened_B_1_1_6561 | 54.7 | 9.521E-132 | 439 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_497514.T0 |
| Description | Glycosyl hydrolase family 85 |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd06547 | GH85_ENGase | - | 61 | 436 |
| Pfam | PF03644 | Glycosyl hydrolase family 85 | IPR005201 | 75 | 416 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR005201 | Glycoside hydrolase, family 85 |
| IPR032979 | Cytosolic endo-beta-N-acetylglucosaminidase |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005737 | cytoplasm | CC |
| GO:0033925 | mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K01227 |
EggNOG
| COG category | Description |
|---|---|
| G | Glycosyl hydrolase family 85 |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH85 |
Transcription factor
| Group |
|---|
| No records |
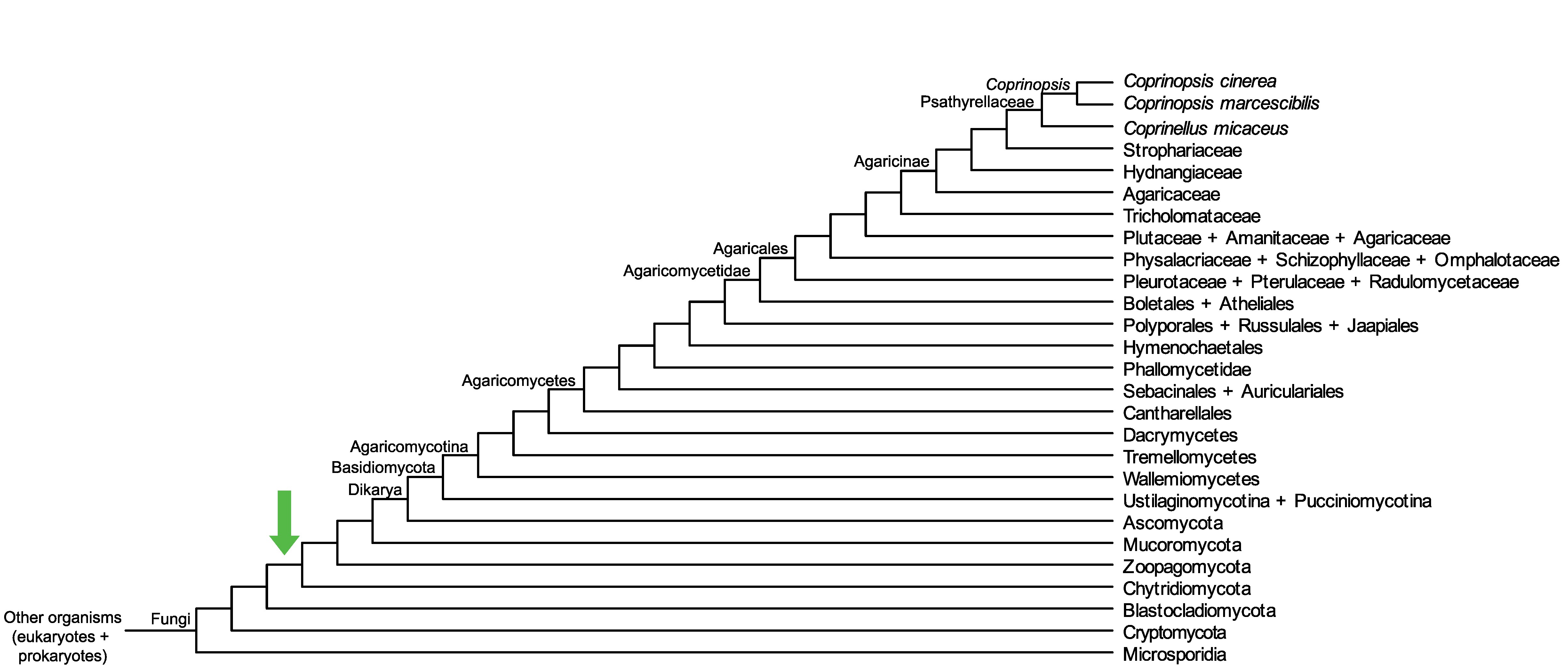
Conservation of CopciAB_497514 across fungi.
Arrow shows the origin of gene family containing CopciAB_497514.
Protein
| Sequence id | CopciAB_497514.T0 |
|---|---|
| Sequence |
>CopciAB_497514.T0 MPIAGKKFHPRALPEFWRTFREMDEWRATQTGPQARPAEGILKYVPRKIRPADIAGKGRLLVSHDYKGGYVEDPF SKSYTFNWWFSTDSFNYFAHHRITIPPPEWINAAHRQGVPILGTIIFEGGSDEDILRMVIGKTPGSTSNFHAERN AEYTVPVSSYYAELFADLAVERGFDGWLLNVEIGLQGGSEQARGLAAWVALLQQEVLKKVGPHGLVIWYDSVTVR GDLWWQDRLNAFNLPFFLNSSGIFTNYWWYNDAPQKQIDFLSRVDPNLTGQTAEPHQYNLQKTIQDIYIGVDVWG RGSHGGGGFGAYKAIEHADPKGLGFSVALFAQGWTWETEEEKPGWNWAQFWDYDSKLWVGPPGVVEAPDHTVKPG EYPCVHGPFQPISSFFLTYPPPDPLDLPFYTNFCPGIGDAWFVEGKEVFRSETGWTDMDKQTTVGDLVWPRPKIY DLPSQNASQATLNAAFNFNDAWNGGNSLQINLSVPGGATTYGAYWVPIQTFTFSSRRQYEASIVYKPGLSGKTRF DAKYEVGIRTVTGEDQGKIISNTTTEIGNGWRKVHILFEIETPIEGGSIIVPSSIGLVIAVSNVSTTEQFEFPFL VGQIAIHPHLPDRYKEFKPALLWLLFTPSAGTNSLDGTLTWDVVAAIERPPPVEINNPDDAQIPWNLQPTKQEWF PDFLYFNVYVLELLDGGGQGPPQWIGTTGYDGEKKRFFIYDESLPPTSGLRRFTFQIEGVLETGESTHWYDAPAA PSATAGGEQKRTRRTSLKSVLSPLRRKKSKGDISVAK |
| Length | 787 |
Coding
| Sequence id | CopciAB_497514.T0 |
|---|---|
| Sequence |
>CopciAB_497514.T0 ATGCCTATCGCTGGGAAGAAGTTCCACCCCCGGGCTCTGCCTGAGTTCTGGAGGACGTTCCGGGAGATGGACGAA TGGCGGGCCACTCAGACTGGACCTCAGGCTCGTCCGGCTGAAGGCATTCTCAAGTACGTTCCACGGAAGATTCGG CCTGCTGATATCGCAGGGAAAGGTCGACTGCTGGTTTCTCATGACTACAAGGGAGGCTACGTCGAAGATCCCTTT TCCAAGTCGTATACCTTCAATTGGTGGTTCTCGACGGATAGCTTCAACTACTTCGCTCACCACCGGATAACCATT CCCCCTCCGGAATGGATAAACGCTGCTCATCGCCAGGGTGTACCTATTCTCGGCACCATCATCTTCGAAGGTGGA AGCGACGAAGACATCCTCCGGATGGTGATCGGGAAAACACCAGGAAGCACCAGCAACTTCCACGCCGAACGAAAC GCGGAGTACACCGTACCAGTTTCGTCGTACTACGCAGAACTCTTCGCAGACCTGGCTGTGGAGCGTGGATTCGAT GGTTGGCTGCTCAACGTTGAGATTGGTCTGCAGGGAGGATCAGAGCAAGCACGAGGTCTGGCTGCTTGGGTTGCG CTACTGCAGCAGGAGGTTTTGAAGAAGGTTGGCCCACATGGCTTGGTCATTTGGTACGACAGTGTCACTGTCCGT GGGGACCTGTGGTGGCAAGACAGGCTGAACGCTTTCAACTTGCCGTTCTTCTTGAATTCTTCGGGAATTTTCACA AACTATTGGTGGTACAACGATGCGCCTCAGAAACAAATCGACTTCCTTTCGAGGGTTGACCCGAATCTCACCGGG CAAACCGCTGAGCCGCATCAATACAACCTGCAGAAGACGATTCAAGATATCTATATCGGTGTGGATGTCTGGGGA CGCGGTTCGCATGGTGGAGGAGGATTTGGTGCCTACAAAGCTATTGAGCACGCAGACCCGAAAGGACTCGGGTTT AGCGTTGCCCTCTTCGCTCAAGGATGGACCTGGGAAACAGAGGAGGAGAAACCAGGCTGGAACTGGGCACAGTTC TGGGACTACGACTCTAAACTCTGGGTTGGACCTCCCGGAGTTGTCGAGGCACCTGACCATACCGTCAAACCTGGC GAATACCCCTGCGTTCACGGACCCTTCCAACCCATCTCCAGCTTCTTCCTGACATATCCACCTCCCGACCCGCTA GACCTACCATTCTACACCAACTTCTGCCCCGGTATCGGAGATGCGTGGTTCGTTGAGGGCAAGGAGGTCTTCCGC TCCGAGACGGGTTGGACAGACATGGACAAGCAGACCACGGTTGGCGACTTGGTCTGGCCCCGACCCAAGATTTAT GATCTTCCATCTCAAAATGCCAGCCAGGCTACGTTAAATGCGGCATTCAACTTTAACGATGCGTGGAATGGAGGG AACTCGCTTCAGATCAACCTCAGCGTCCCTGGAGGGGCGACCACGTATGGAGCCTACTGGGTTCCCATCCAGACA TTCACATTCTCCAGTCGGCGCCAGTACGAAGCTTCGATCGTCTACAAGCCGGGATTGAGTGGGAAGACTCGGTTC GATGCCAAGTACGAGGTGGGTATCCGAACTGTTACAGGGGAAGACCAAGGGAAGATCATCTCCAACACGACTACG GAAATTGGAAATGGTTGGCGCAAAGTGCACATCCTATTCGAGATCGAGACCCCAATTGAAGGAGGATCCATCATC GTACCTTCATCCATCGGTCTGGTCATTGCAGTTTCGAACGTATCAACTACCGAGCAATTCGAGTTCCCCTTCCTG GTCGGCCAGATCGCCATCCACCCCCACCTCCCCGATCGTTACAAGGAGTTCAAGCCGGCCCTCCTGTGGCTTTTG TTCACACCTTCCGCTGGAACTAATAGCCTCGATGGCACCCTCACTTGGGACGTCGTCGCAGCCATTGAACGCCCT CCGCCAGTCGAAATTAACAACCCCGATGACGCACAAATCCCCTGGAACCTGCAGCCGACCAAACAAGAATGGTTC CCCGACTTCCTCTACTTCAATGTGTACGTGCTGGAGCTCTTGGATGGTGGTGGACAAGGTCCCCCACAGTGGATT GGCACGACTGGATACGATGGGGAGAAGAAGAGGTTCTTCATCTATGACGAAAGCTTGCCACCGACGTCCGGTTTA AGGAGGTTCACGTTCCAAATCGAGGGCGTCCTGGAGACGGGAGAGTCAACGCACTGGTACGATGCACCGGCTGCG CCGTCGGCAACTGCAGGAGGAGAGCAGAAGAGGACACGACGCACCTCTCTCAAGTCCGTGCTCAGTCCGTTGCGG AGGAAAAAGTCGAAGGGGGATATCTCCGTCGCTAAGTGA |
| Length | 2364 |
Transcript
| Sequence id | CopciAB_497514.T0 |
|---|---|
| Sequence |
>CopciAB_497514.T0 ATGTTATCATACCAGCTGCCACGACCCCTCTCTCCAGTACCCAACCACCATGCCTATCGCTGGGAAGAAGTTCCA CCCCCGGGCTCTGCCTGAGTTCTGGAGGACGTTCCGGGAGATGGACGAATGGCGGGCCACTCAGACTGGACCTCA GGCTCGTCCGGCTGAAGGCATTCTCAAGTACGTTCCACGGAAGATTCGGCCTGCTGATATCGCAGGGAAAGGTCG ACTGCTGGTTTCTCATGACTACAAGGGAGGCTACGTCGAAGATCCCTTTTCCAAGTCGTATACCTTCAATTGGTG GTTCTCGACGGATAGCTTCAACTACTTCGCTCACCACCGGATAACCATTCCCCCTCCGGAATGGATAAACGCTGC TCATCGCCAGGGTGTACCTATTCTCGGCACCATCATCTTCGAAGGTGGAAGCGACGAAGACATCCTCCGGATGGT GATCGGGAAAACACCAGGAAGCACCAGCAACTTCCACGCCGAACGAAACGCGGAGTACACCGTACCAGTTTCGTC GTACTACGCAGAACTCTTCGCAGACCTGGCTGTGGAGCGTGGATTCGATGGTTGGCTGCTCAACGTTGAGATTGG TCTGCAGGGAGGATCAGAGCAAGCACGAGGTCTGGCTGCTTGGGTTGCGCTACTGCAGCAGGAGGTTTTGAAGAA GGTTGGCCCACATGGCTTGGTCATTTGGTACGACAGTGTCACTGTCCGTGGGGACCTGTGGTGGCAAGACAGGCT GAACGCTTTCAACTTGCCGTTCTTCTTGAATTCTTCGGGAATTTTCACAAACTATTGGTGGTACAACGATGCGCC TCAGAAACAAATCGACTTCCTTTCGAGGGTTGACCCGAATCTCACCGGGCAAACCGCTGAGCCGCATCAATACAA CCTGCAGAAGACGATTCAAGATATCTATATCGGTGTGGATGTCTGGGGACGCGGTTCGCATGGTGGAGGAGGATT TGGTGCCTACAAAGCTATTGAGCACGCAGACCCGAAAGGACTCGGGTTTAGCGTTGCCCTCTTCGCTCAAGGATG GACCTGGGAAACAGAGGAGGAGAAACCAGGCTGGAACTGGGCACAGTTCTGGGACTACGACTCTAAACTCTGGGT TGGACCTCCCGGAGTTGTCGAGGCACCTGACCATACCGTCAAACCTGGCGAATACCCCTGCGTTCACGGACCCTT CCAACCCATCTCCAGCTTCTTCCTGACATATCCACCTCCCGACCCGCTAGACCTACCATTCTACACCAACTTCTG CCCCGGTATCGGAGATGCGTGGTTCGTTGAGGGCAAGGAGGTCTTCCGCTCCGAGACGGGTTGGACAGACATGGA CAAGCAGACCACGGTTGGCGACTTGGTCTGGCCCCGACCCAAGATTTATGATCTTCCATCTCAAAATGCCAGCCA GGCTACGTTAAATGCGGCATTCAACTTTAACGATGCGTGGAATGGAGGGAACTCGCTTCAGATCAACCTCAGCGT CCCTGGAGGGGCGACCACGTATGGAGCCTACTGGGTTCCCATCCAGACATTCACATTCTCCAGTCGGCGCCAGTA CGAAGCTTCGATCGTCTACAAGCCGGGATTGAGTGGGAAGACTCGGTTCGATGCCAAGTACGAGGTGGGTATCCG AACTGTTACAGGGGAAGACCAAGGGAAGATCATCTCCAACACGACTACGGAAATTGGAAATGGTTGGCGCAAAGT GCACATCCTATTCGAGATCGAGACCCCAATTGAAGGAGGATCCATCATCGTACCTTCATCCATCGGTCTGGTCAT TGCAGTTTCGAACGTATCAACTACCGAGCAATTCGAGTTCCCCTTCCTGGTCGGCCAGATCGCCATCCACCCCCA CCTCCCCGATCGTTACAAGGAGTTCAAGCCGGCCCTCCTGTGGCTTTTGTTCACACCTTCCGCTGGAACTAATAG CCTCGATGGCACCCTCACTTGGGACGTCGTCGCAGCCATTGAACGCCCTCCGCCAGTCGAAATTAACAACCCCGA TGACGCACAAATCCCCTGGAACCTGCAGCCGACCAAACAAGAATGGTTCCCCGACTTCCTCTACTTCAATGTGTA CGTGCTGGAGCTCTTGGATGGTGGTGGACAAGGTCCCCCACAGTGGATTGGCACGACTGGATACGATGGGGAGAA GAAGAGGTTCTTCATCTATGACGAAAGCTTGCCACCGACGTCCGGTTTAAGGAGGTTCACGTTCCAAATCGAGGG CGTCCTGGAGACGGGAGAGTCAACGCACTGGTACGATGCACCGGCTGCGCCGTCGGCAACTGCAGGAGGAGAGCA GAAGAGGACACGACGCACCTCTCTCAAGTCCGTGCTCAGTCCGTTGCGGAGGAAAAAGTCGAAGGGGGATATCTC CGTCGCTAAGTGAGAGAAAGAGAGCGAGAAAAAGCAGAAATGTATAAATAGTCCTTTGTGTTGTACAGATGCTGA ACCATGGATTACTTGTCCGATCCTACCAGTCCGCGAATCCTACCAATTTGCCATCCC |
| Length | 2532 |
Gene
| Sequence id | CopciAB_497514.T0 |
|---|---|
| Sequence |
>CopciAB_497514.T0 ATGTTATCATACCAGCTGCCACGACCCCTCTCTCCAGTACCCAACCACCATGCCTATCGCTGGGAAGAAGTTCCA CCCCCGGGCTCTGCCTGAGTTCTGGAGGACGTTCCGGGAGATGGACGAATGGCGGGCCACTCAGACTGGACCTCA GGCTCGTCCGGCTGAAGGCATTCTCAAGTACGTTCCACGGAAGATTCGGCCTGCTGATATCGCAGGGAAAGGTCG ACTGCTGGTAAGCGCCCACTGCTTCAGGTACTCTTCCGACTCTGGGCTCATTGGAAGCATTTCAGGTTTCTCATG ACTACAAGGGAGGCTACGTCGAAGATCCCTTTTCCAAGTCGTATACCTTCAATTGGTGGTTCTCGACGGATAGCT TCAACTAGTGAGCCATACAATTACCTGTGTCCCTGGAGGTTGACGAACAATCCCAAAGCTTCGCTCACCACCGGA TAACCATTCCCCCTCCGGAATGGATAAACGCTGCTCATCGCCAGGGTGTACCTATTCTCGGCACCATGTACGTGA TACTCGATCCAGAAACCTTTCTTCACACTGACCATGCACAAGCATCTTCGAAGGTGGAAGCGACGAAGACATCCT CCGGATGGTGATCGGGAAAACACCAGGAAGCACCAGCAACTTCCACGCCGAACGAAACGCGGAGTACACCGTACC AGTTTCGTCGTACTACGCAGAACTCTTCGCAGACCTGGCTGTGGAGCGTGGATTCGATGGTTGGCTGCTCAACGT TGAGATTGGTCTGCAGGGAGGATCAGAGCAAGCACGAGGTCTGGCTGCTTGGGTTGCGCTACTGCAGCAGGAGGT TTTGAAGAAGGTTGGCCCACATGGCTTGGTCATTTGGTACGACAGTGTCACTGTCCGTGGGGACCTGTGGTGGCA AGACAGGCTGAACGCTTTCAACTTGCCGTTCTTCTTGAATTCTTCGGGAATTTTCACAAACTATTGGGTATGTGG CTGATTTGTTGTTCCTTTGACTCCAACTGACTGGCACTTTCCCAGTGGTACAACGATGCGCCTCAGAAACAAATC GACTTCCTTTCGAGGGTTGACCCGAATCTCACCGGGCAAACCGCTGAGCCGCATCAATACAACCTGCAGAAGACG ATTCAAGATATCTATATCGGTGTGGATGTCTGGGGACGCGGTTCGCATGGTGGAGGAGGATTTGGTGCCTACAAA GCTATTGAGCACGCAGACCCGAAAGGACTCGGGTTTAGCGTTGCCCTCTTCGCTCAAGGATGGACCTGGGAAACA GAGGAGGAGAAACCAGGCTGGAACTGGGCACAGTTCTGGGACTACGACTCTAAACTCTGGGTTGGACCTCCCGGA GTTGTCGAGGCACCTGACCATACCGTCAAACCTGGCGAATACCCCTGCGTTCACGGACCCTTCCAACCCATCTCC AGCTTCTTCCTGACATATCCACCTCCCGACCCGCTAGACCTACCATTCTACACCAACTTCTGCCCCGGTATCGGA GATGCGTGGTTCGTTGAGGGCAAGGAGGTCTTCCGCTCCGAGACGGGTTGGACAGACATGGACAAGCAGACCACG GTTGGCGACTTGGTCTGGCCCCGACCCAAGATTTATGATCTTCCATCTCAAAATGCCAGCCAGGCTACGTTAAAT GCGGCATTCAACTTTAACGATGCGTGGAATGGAGGGAACTCGCTTCAGATCAACCTCAGCGTCCCTGGAGGGGCG ACCACGTATGGAGCCTACTGGGTTCCCATCCAGACATTCACATTCTCCAGTCGGCGCCAGTACGAAGCTTCGATC GTCTACAAGCCGGGATTGAGTGGGAAGACTCGGTTCGATGCCAAGTACGAGGTGGGTATCCGAACTGTTACAGGG GAAGACCAAGGGAAGATCATCTCCAACACGACTACGGAAATTGGAAATGGTTGGCGCAAAGTGCACATCCTATTC GAGATCGAGACCCCAATTGAAGGAGGATCCATCATCGTACCTTCATCCATCGGTCTGGTCATTGCAGTTTCGAAC GTATCAACTACCGAGCAATTCGAGTTCCCCTTCCTGGTCGGCCAGATCGCCATCCACCCCCACCTCCCCGATCGT TACAAGGAGTTCAAGCCGGCCCTCCTGTGGCTTTTGTTCACACCTTCCGCTGGAACTAATAGCCTCGATGGCACC CTCACTTGGGACGTCGTCGCAGCCATTGAACGCCCTCCGCCAGTCGAAATTAACAACCCCGATGACGCACAAATC CCCTGGAACCTGCAGCCGACCAAACAAGAATGGTTCCCCGACTTCCTCTACTTCAATGTGTACGTGCTGGAGCTC TTGGATGGTGGTGGACAAGGTCCCCCACAGTGGATTGGCACGACTGGATACGATGGGGAGAAGAAGAGGTTCTTC ATCTATGACGAAAGCTTGCCACCGACGTCCGGTTTAAGGAGGTTCACGTTCCAAATCGAGGGCGTCCTGGAGACG GGAGAGTCAACGCACTGGTACGATGCACCGGCTGCGCCGTCGGCAACTGCAGGAGGAGAGCAGAAGAGGACACGA CGCACCTCTCTCAAGTCCGTGCTCAGTCCGTTGCGGAGGAAAAAGTCGAAGGGGGATATCTCCGTCGCTAAGTGA GAGAAAGAGAGCGAGAAAAAGCAGAAATGTATAAATAGTCCTTTGTGTTGTACAGATGCTGAACCATGGATTACT TGTCCGATCCTACCAGTCCGCGAATCCTACCAATTTGCCATCCC |
| Length | 2744 |