CopciAB_497939
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_497939 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 497939 |
| Uniprot id | Functional description | Cytochrome domain of cellobiose dehydrogenase | |
| Location | scaffold_11:1310014..1313628 | Strand | + |
| Gene length (nt) | 3615 | Transcript length (nt) | 2741 |
| CDS length (nt) | 2379 | Protein length (aa) | 792 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN7230 |
| Neurospora crassa | cdh-2 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7262555 | 65.7 | 0 | 1092 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB26844 | 65.7 | 0 | 1082 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1592473 | 63.5 | 0 | 1030 |
| Pleurotus ostreatus PC15 | PleosPC15_2_41743 | 64 | 0 | 1016 |
| Pleurotus ostreatus PC9 | PleosPC9_1_62103 | 64 | 0 | 1016 |
| Schizophyllum commune H4-8 | Schco3_2642438 | 62.8 | 0 | 1005 |
| Flammulina velutipes | Flave_chr01AA00302 | 61.5 | 1.976E-304 | 997 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_86428 | 60.6 | 5.958E-304 | 987 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_6040 | 60.7 | 6.442E-304 | 987 |
| Lentinula edodes NBRC 111202 | Lenedo1_1176330 | 59.4 | 1.852E-304 | 977 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_107890 | 57.2 | 7.952E-304 | 943 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_188178 | 56.9 | 1.69E-304 | 942 |
| Auricularia subglabra | Aurde3_1_1329463 | 55.2 | 4.273E-283 | 881 |
| Lentinula edodes B17 | Lened_B_1_1_5826 | 58.1 | 5.532E-271 | 845 |
| Grifola frondosa | Grifr_OBZ78256 | 44.8 | 1.758E-205 | 655 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_497939.T0 |
| Description | Cytochrome domain of cellobiose dehydrogenase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd09630 | CDH_like_cytochrome | IPR015920 | 46 | 210 |
| Pfam | PF16010 | Cytochrome domain of cellobiose dehydrogenase | IPR015920 | 47 | 223 |
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | 259 | 532 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | 658 | 780 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 42 | 0.8112 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal |
| IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal |
| IPR015920 | Cellobiose dehydrogenase, cytochrome domain |
| IPR036188 | FAD/NAD(P)-binding domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF |
| GO:0050660 | flavin adenine dinucleotide binding | MF |
KEGG
| KEGG Orthology |
|---|
| K19069 |
EggNOG
| COG category | Description |
|---|---|
| E | Cytochrome domain of cellobiose dehydrogenase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA8 | |
| AA | AA3 | AA3_1 |
Transcription factor
| Group |
|---|
| No records |
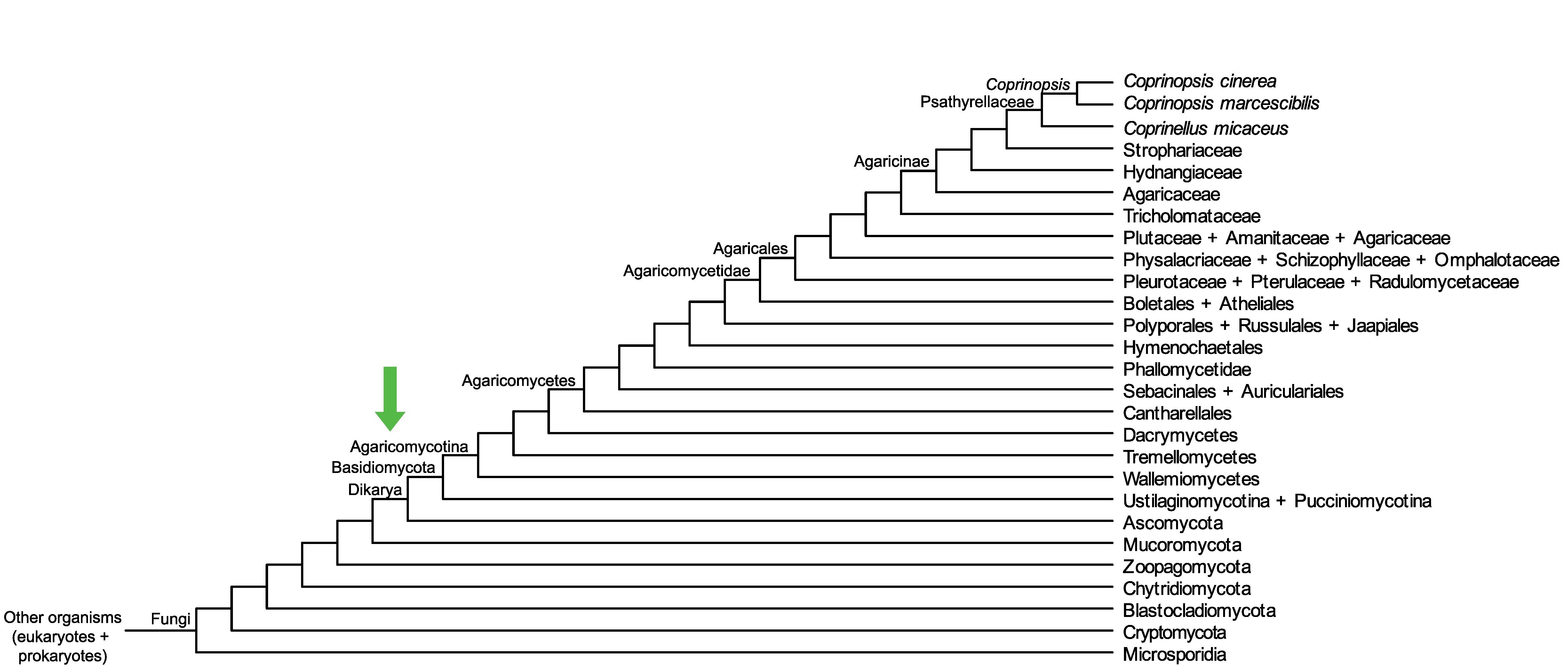
Conservation of CopciAB_497939 across fungi.
Arrow shows the origin of gene family containing CopciAB_497939.
Protein
| Sequence id | CopciAB_497939.T0 |
|---|---|
| Sequence |
>CopciAB_497939.T0 MYKKNGFRLVHVPHRFSPSQPTRTTMLGRLFTSLLLAGLALAQTESYVDPDTGITFQGRTDPVHGVTIGYVLPPL EPASDEFIGQILAPIENGWVGIAPGGGMINNLLVVAWPNGNEVVASVRMANTYVLPQPFNDPVLTILPSTKVNAT HWKLDYRCQGCTTWETANGPRSLPIDSAGAAAWALSKSPVDDPSDPDTTFAQHTDFGFYGQIWALSHVDAETYEH WASGGTGGGPTPTTPPTEPPTPTEPVPTVEPTPYDYIVVGAGPGGLVTADRLSEAGHKVLLIERGGPSLGSTGGT TQPDWLKGTNLTKFDVPGLFETMFMDADPYWWCKDINVFAGCLLGGGTAINGGLYWYPPDIDFSHTYGWPPSWQS HHQYTDKLKQRIPSTDAPSTDGKRYLMETFDVVEKLLRPQGYHQQTINNNPNQKDRVYGYSAYSFIDGKRAGPIA TYYKTAIARPNLTYFSHTTVLNVVREGSTITGVNTDNPLIGPNGFVPLNPRGRVILSAGAFGSARLLFRSGIGPR DMLEIVKNDRNAGPNMPAEEDWIDLPVGENVQDNPSINLVFTHPDIDAYENWANVWSNPRQADAAQYIDSQSGVF SQSSPRINFWRKLMGSDKKPRWMQGTARPGAASITTDFPYNASNIFTITTYLATGITSRGRIGIDAALTARALKN PWFTDPVDKEVLLKGIHELIDAMEHVPGLTLITPDNTTTIEDYVDNYPTGWLNSNHWVGSNSIGKVVDENTKVFN TDNLFVVDASIIPSMPMGNPQGAIMSTAEQGVARILALQGGP |
| Length | 792 |
Coding
| Sequence id | CopciAB_497939.T0 |
|---|---|
| Sequence |
>CopciAB_497939.T0 ATGTATAAAAAGAATGGATTTAGGCTTGTTCACGTTCCTCACCGCTTTTCCCCTTCCCAACCCACACGAACGACG ATGTTGGGACGACTCTTTACGAGTCTCCTCCTCGCAGGGCTCGCCCTCGCGCAGACTGAATCGTATGTCGACCCT GACACCGGGATCACCTTCCAAGGTAGAACCGACCCCGTGCACGGCGTGACCATTGGTTACGTCCTTCCACCGCTC GAGCCTGCCTCCGATGAATTCATTGGCCAAATTCTTGCGCCCATTGAGAACGGATGGGTCGGCATTGCCCCTGGT GGAGGCATGATTAACAACCTCCTCGTCGTTGCTTGGCCTAATGGCAACGAGGTCGTTGCTTCCGTTCGGATGGCA AACACCTACGTCCTCCCTCAACCTTTCAACGATCCTGTCCTTACCATTCTTCCATCGACCAAGGTCAACGCGACC CACTGGAAACTCGACTACCGCTGCCAAGGCTGCACCACTTGGGAAACCGCCAACGGACCCCGCTCACTCCCCATC GACTCTGCCGGTGCCGCCGCCTGGGCACTCTCCAAGTCCCCCGTCGACGACCCCTCCGACCCCGACACCACCTTC GCTCAACACACTGACTTCGGCTTCTACGGCCAAATCTGGGCTCTCTCCCATGTTGACGCTGAGACCTACGAGCAC TGGGCTTCTGGAGGCACTGGTGGCGGTCCCACCCCTACCACTCCTCCCACTGAACCTCCTACTCCTACTGAGCCT GTTCCTACCGTAGAGCCTACTCCCTACGACTACATCGTCGTCGGAGCCGGCCCCGGTGGATTGGTCACTGCTGAC CGTCTCTCCGAGGCAGGCCACAAGGTCCTCCTTATTGAGAGGGGTGGACCCAGCTTGGGCTCGACCGGCGGTACC ACTCAGCCCGATTGGTTGAAGGGAACTAACCTCACGAAATTCGATGTCCCTGGATTGTTCGAGACTATGTTCATG GATGCCGATCCTTACTGGTGGTGCAAGGACATCAACGTTTTCGCCGGATGCTTGCTCGGCGGAGGAACTGCTATC AACGGAGGCTTGTACTGGTACCCTCCCGACATCGATTTCTCTCACACCTACGGCTGGCCTCCCTCGTGGCAGAGC CATCACCAATACACCGACAAGCTTAAGCAGCGTATCCCCAGTACTGATGCTCCCTCGACCGATGGCAAGCGATAT CTCATGGAGACCTTCGACGTCGTCGAGAAGTTGCTCCGCCCTCAAGGATACCACCAGCAAACCATCAACAACAAC CCTAACCAGAAGGATAGGGTTTACGGTTACAGCGCCTACTCTTTCATCGACGGCAAGCGCGCTGGTCCCATCGCA ACCTACTACAAGACCGCCATCGCCCGACCCAACTTGACCTACTTCTCGCACACCACCGTCCTCAACGTCGTTCGT GAAGGCTCCACCATCACCGGTGTCAACACCGACAACCCATTGATCGGTCCCAACGGCTTCGTCCCTCTCAACCCC CGCGGCCGTGTCATCCTCTCCGCTGGCGCCTTCGGCTCTGCACGTCTCCTCTTCCGCTCTGGAATTGGCCCTCGC GATATGCTCGAGATCGTCAAGAATGACAGGAACGCCGGCCCCAACATGCCCGCTGAGGAGGACTGGATCGACCTC CCTGTTGGCGAGAACGTCCAGGACAACCCTTCGATTAACTTGGTCTTCACCCACCCCGACATTGATGCCTATGAG AACTGGGCCAACGTTTGGAGCAACCCCAGGCAGGCTGATGCTGCTCAGTATATCGACAGCCAGTCTGGCGTCTTC TCTCAATCTTCCCCTCGTATCAACTTCTGGAGGAAACTTATGGGCAGCGACAAGAAGCCTCGATGGATGCAAGGA ACTGCTCGCCCTGGTGCCGCCTCCATCACCACCGACTTCCCCTACAACGCTAGCAACATCTTCACCATCACCACC TACCTCGCCACTGGTATCACCTCCCGTGGTCGTATTGGTATCGATGCTGCCCTTACTGCTCGTGCCCTCAAGAAC CCCTGGTTCACCGACCCCGTCGACAAAGAGGTCCTCCTCAAGGGTATCCATGAACTCATTGACGCCATGGAGCAT GTCCCCGGATTGACCCTCATCACTCCCGACAACACGACCACCATCGAGGACTACGTCGACAACTACCCCACCGGC TGGCTCAACTCCAACCACTGGGTCGGGTCCAACTCCATCGGAAAGGTTGTCGATGAGAACACCAAGGTCTTCAAC ACCGATAACCTCTTCGTTGTTGATGCTTCCATCATTCCTTCGATGCCCATGGGTAACCCACAGGGTGCTATCATG TCGACTGCTGAACAGGGTGTGGCTAGGATTCTCGCCCTCCAAGGCGGCCCTTAG |
| Length | 2379 |
Transcript
| Sequence id | CopciAB_497939.T0 |
|---|---|
| Sequence |
>CopciAB_497939.T0 TCGGGTCTACAGGACGATCGGTCATGAGAGCCATCATCACAACCGAATTGCCACCGTTCCTATTGGCCTACGACT CCATCGCACTCGCAATAGCTTCTATGTCGACTCGTATTCCCAGTACCCATGACGCGGACGAATGTATATCTTCCA GCCAACCGTTGTGTGGGCATGGGTTTTTGAACACCTGAAGGGGAAATAAATCATTTACACCTTCTGTCTCCACCT GGACACGCGGAACTATCTCGCCATTTAGACAGTATGCGGTACGCAGAGGGTGGATGTATAAAAAGAATGGATTTA GGCTTGTTCACGTTCCTCACCGCTTTTCCCCTTCCCAACCCACACGAACGACGATGTTGGGACGACTCTTTACGA GTCTCCTCCTCGCAGGGCTCGCCCTCGCGCAGACTGAATCGTATGTCGACCCTGACACCGGGATCACCTTCCAAG GTAGAACCGACCCCGTGCACGGCGTGACCATTGGTTACGTCCTTCCACCGCTCGAGCCTGCCTCCGATGAATTCA TTGGCCAAATTCTTGCGCCCATTGAGAACGGATGGGTCGGCATTGCCCCTGGTGGAGGCATGATTAACAACCTCC TCGTCGTTGCTTGGCCTAATGGCAACGAGGTCGTTGCTTCCGTTCGGATGGCAAACACCTACGTCCTCCCTCAAC CTTTCAACGATCCTGTCCTTACCATTCTTCCATCGACCAAGGTCAACGCGACCCACTGGAAACTCGACTACCGCT GCCAAGGCTGCACCACTTGGGAAACCGCCAACGGACCCCGCTCACTCCCCATCGACTCTGCCGGTGCCGCCGCCT GGGCACTCTCCAAGTCCCCCGTCGACGACCCCTCCGACCCCGACACCACCTTCGCTCAACACACTGACTTCGGCT TCTACGGCCAAATCTGGGCTCTCTCCCATGTTGACGCTGAGACCTACGAGCACTGGGCTTCTGGAGGCACTGGTG GCGGTCCCACCCCTACCACTCCTCCCACTGAACCTCCTACTCCTACTGAGCCTGTTCCTACCGTAGAGCCTACTC CCTACGACTACATCGTCGTCGGAGCCGGCCCCGGTGGATTGGTCACTGCTGACCGTCTCTCCGAGGCAGGCCACA AGGTCCTCCTTATTGAGAGGGGTGGACCCAGCTTGGGCTCGACCGGCGGTACCACTCAGCCCGATTGGTTGAAGG GAACTAACCTCACGAAATTCGATGTCCCTGGATTGTTCGAGACTATGTTCATGGATGCCGATCCTTACTGGTGGT GCAAGGACATCAACGTTTTCGCCGGATGCTTGCTCGGCGGAGGAACTGCTATCAACGGAGGCTTGTACTGGTACC CTCCCGACATCGATTTCTCTCACACCTACGGCTGGCCTCCCTCGTGGCAGAGCCATCACCAATACACCGACAAGC TTAAGCAGCGTATCCCCAGTACTGATGCTCCCTCGACCGATGGCAAGCGATATCTCATGGAGACCTTCGACGTCG TCGAGAAGTTGCTCCGCCCTCAAGGATACCACCAGCAAACCATCAACAACAACCCTAACCAGAAGGATAGGGTTT ACGGTTACAGCGCCTACTCTTTCATCGACGGCAAGCGCGCTGGTCCCATCGCAACCTACTACAAGACCGCCATCG CCCGACCCAACTTGACCTACTTCTCGCACACCACCGTCCTCAACGTCGTTCGTGAAGGCTCCACCATCACCGGTG TCAACACCGACAACCCATTGATCGGTCCCAACGGCTTCGTCCCTCTCAACCCCCGCGGCCGTGTCATCCTCTCCG CTGGCGCCTTCGGCTCTGCACGTCTCCTCTTCCGCTCTGGAATTGGCCCTCGCGATATGCTCGAGATCGTCAAGA ATGACAGGAACGCCGGCCCCAACATGCCCGCTGAGGAGGACTGGATCGACCTCCCTGTTGGCGAGAACGTCCAGG ACAACCCTTCGATTAACTTGGTCTTCACCCACCCCGACATTGATGCCTATGAGAACTGGGCCAACGTTTGGAGCA ACCCCAGGCAGGCTGATGCTGCTCAGTATATCGACAGCCAGTCTGGCGTCTTCTCTCAATCTTCCCCTCGTATCA ACTTCTGGAGGAAACTTATGGGCAGCGACAAGAAGCCTCGATGGATGCAAGGAACTGCTCGCCCTGGTGCCGCCT CCATCACCACCGACTTCCCCTACAACGCTAGCAACATCTTCACCATCACCACCTACCTCGCCACTGGTATCACCT CCCGTGGTCGTATTGGTATCGATGCTGCCCTTACTGCTCGTGCCCTCAAGAACCCCTGGTTCACCGACCCCGTCG ACAAAGAGGTCCTCCTCAAGGGTATCCATGAACTCATTGACGCCATGGAGCATGTCCCCGGATTGACCCTCATCA CTCCCGACAACACGACCACCATCGAGGACTACGTCGACAACTACCCCACCGGCTGGCTCAACTCCAACCACTGGG TCGGGTCCAACTCCATCGGAAAGGTTGTCGATGAGAACACCAAGGTCTTCAACACCGATAACCTCTTCGTTGTTG ATGCTTCCATCATTCCTTCGATGCCCATGGGTAACCCACAGGGTGCTATCATGTCGACTGCTGAACAGGGTGTGG CTAGGATTCTCGCCCTCCAAGGCGGCCCTTAGACACGGCGAAGTATTTATTTTATAAAGGAAGGGGAATTAACAT AGTTGCCGTAGCTACCTCAATATAAAAAAGGTTCATAATTC |
| Length | 2741 |
Gene
| Sequence id | CopciAB_497939.T0 |
|---|---|
| Sequence |
>CopciAB_497939.T0 TCGGGTCTACAGGACGATCGGTCATGAGAGCCATCATCACAACCGAATTGCCACCGTTCCTATTGGCCTACGACT CCATCGCACTCGCAATAGCTTCTATGTCGACTCGTATTCCCAGTACCCATGACGCGGACGAATGTATATCTTCCA GCCAACCGTTGTGTGGGCATGGGTTTTTGAACACCTGAAGGGGAAATAAATCATTTACACCTTCTGTCTCCACCT GGACACGCGGAACTATCTCGCCATTTAGACAGTATGCGGTACGCAGAGGGTGGATGTATAAAAAGAATGGATTTA GGCTTGTTCACGTTCCTCACCGCTTTTCCCCTTCCCAACCCACACGAACGACGATGTTGGGACGACTCTTTACGA GTCTCCTCCTCGCAGGGCTCGGTGCGTCTCATCGTCTTCATCGCACCATATTTAGAAACTAATAGTACCTTCACT AGCCCTCGCGCAGACTGAATCGTATGTCGACCCTGACACCGGGATCACCTTCCAAGGTAGAACCGACCCCGTGCA CGGCGTGACCATTGGTTACGTCCTTCCACCGCTCGAGCCTGCCTCCGATGAATTCATTGGCCAAATTCTTGCGCC CATTGAGAACGGATGGGTCGGCATTGCCCCTGGTGGAGGCATGATTAACAACCTCCTCGTCGTTGCTTGGCCTAA TGGCAACGAGGTCGTTGCTTCCGTTCGGATGGCAAAGTGAGTGCACGTCTTGTGTATAACGCCCCTTGTGGAACG GAGTGCTGACCCATGGTTTTATCTTTCTATAGCACCTACGTCCTCCCTCAGTACGTTACTCTATCAGCGCTTGCT ATCTTCTACCTCGGTCCTCAGACTTATCTCCTACTCTAGACCTTTCAACGATCCTGTCCTTACCATTCTTCCATC GACCAAGGTCAACGCGACCCACTGGAAACTCGACTACCGCTGCCAAGGCTGCACCAGTATGTACCCCCGCCTTCC AATCACACCCAACAGGTCCCTTACCACCATTTCCTTACAGCTTGGGAAACCGCCAACGGACCCCGCTCACTCCCC ATCGACTCTGCCGGTGCCGCCGCCTGGGCACTCTCCAAGTCCCCCGTCGACGACCCCTCCGACCCCGACACCACC TTCGCTCAACACACTGACTGTACGCCTTCTTCCCCGCTCTACCGTTACCCTAGGACACTGCCTAACACTCCTTTG TAGTCGGCTTCTACGGCCAAATCTGGGCTCTCTCCCATGTTGACGCTGAGACCTACGAGCACTGGGCTTCTGGAG GCACTGGTGGCGGTCCCACCCCTACCACTCCTCCCACTGAACCTCCTACTCCTACTGAGCCTGTTCCTACCGTAG AGGTACGCATGTTTTCCTCGACTTCTGACTGCAAATGTTAAACGTGTCTTTTCCTATAGCCTACTCCCTACGACT ACATCGTCGTCGGAGCCGGCCCCGGTGGATTGGTCACTGCTGACCGTCTCTCCGAGGCAGGCCACAAGGTCCTCC TTATTGAGAGGGGTGGACCCAGCTTGGGCTCGACCGGCGGTACCACTCAGCCCGATTGGTTGAAGGGAACTAACG TACGTGATTTATACTTGAATCGGGAAAGCAACACCGTACTGAGAAATGCATTCAGCTCACGAAATTCGATGTCCC TGGATTGTTCGAGACTATGTTCATGGATGCCGATCCTTACTGGTGGTGCAAGGGTACGTTTTGGACATTGCAGGG CTGTGGAGTTGTGGGTTAACCCGGCTATGGACAGACATCAACGTTTTCGCCGGATGCTTGCTCGGCGGAGGAACT GCTATCAACGGAGGGTATGTCGCAGTGGCGTCCTTTCTCGCAAATTATCCCCTGACTAACCACCCAATAGCTTGT ACTGGTACCCTCCCGACATCGATTTCTCTCACACCTACGGCTGGCCTCCCTCGTGGCAGAGCCATCACCAATACA CCGACAAGCTTAAGCAGCGTATCCCCAGTACTGATGCTCCCTCGACCGATGGCAAGCGATATCTCATGGAGACCT TCGACGTCGTCGAGAAGTTGCTCCGCCCTCAAGGATACCACCAGCAAACCATCAACAACAACCCTAACCAGAAGG ATAGGGTTTACGGTTACAGCGCCTACTCTGTGAGTTCCTTGTCTCCATCTCCTCCACTTCCATAACTAACAAACC ACCCCCGTAGTTCATCGACGGCAAGCGCGCTGGTCCCATCGCAACCTACTACAAGACCGCCATCGCCCGACCCAA CTTGACCTACTTCTCGCACACCACCGTCCTCAACGTCGTTCGTGAAGGCTCCACCATCACCGGTGTCAACACCGA CAACCCATTGATCGGTCCCAACGGCTTCGTCCCTCTCAACCCCCGCGGCCGTGTCATCCTCTCCGCTGGCGCCTT CGGCTCTGCACGTCTCCTCTTCCGCTCTGGAATTGGCCCTCGCGATATGCTCGAGATCGTCAAGAATGACAGGAA CGCCGGCCCCAACATGCCCGCTGAGGAGGACTGGATCGACCTCCCTGTTGGCGAGAACGTCCAGGACAACCCTTC GATTAACTTGGTCTTCACCCACCCCGACATTGATGCCTATGAGAACTGGGCCAACGTTTGGAGCAACCCCAGGCA GGCTGATGCTGCTCAGTATATCGACAGCCAGTCTGGCGTCTTCTCTCAATCTTCCCCTCGGTGGGTTTTAATCTT CTGCCTGTGCCTGAACGAGTAACAAACTGACGAAATCTCGTTCGCTAGTATCAACTTCTGGAGGAAACTTATGGG CAGCGACAAGAAGCCTCGATGGGTATGTGACAATAGCGTGGGTCAGGACACCAAAATCTTAACGCCCTTCGTAGA TGCAAGGAACTGCTCGCCCTGGTGCCGCCTCCATCACCACCGACTTCCCCTACAACGCTAGCAACATCTTCACCA TCACCACCTACCTGTACGTGTCAACTCTTCAACCCTTTCTAGACCCTCGCTCTGACCTTGAACCATTTCAGCGCC ACTGGTATCACCTCCCGTGGTCGTATTGGTATCGATGCTGCCCTTACTGCTCGTGCCCTCAAGAACCCCTGGTTC ACCGACCCCGTCGACAAAGAGGTCCTCCTCAAGGGTATCCATGAACTCATTGACGCCATGGAGCATGGTACGTAC CAGTGCTTCGTCTTACCGACTTTGACCTGAGGTTCTGTCCCACCAGTCCCCGGATTGACCCTCATCACTCCCGAC AACACGACCACCATCGAGGACTACGTCGACAACTACCCCACCGGCTGGCTCAACTCCAACCACTGGGTCGGGTCC AACTCCATCGGAAAGGTTGTCGATGAGAACACCAAGGTCTTCAACACCGATAACCTCTTCGTTGTTGATGCTTCC ATCGTACGTCGCTTGTCATATTTTCTCAATACCAAAGGTTTTCTGACTTGTGCGATCCAGATTCCTTCGATGCCC ATGGGTAACCCACAGGGTGCTATCATGTCGACTGCTGAACAGGGTGTGGCTAGGATTCTCGCCCTCCAAGGCGGC CCTTAGACACGGCGAAGTATTTATTTTATAAAGGAAGGGGAATTAACATAGTTGCCGTAGCTACCTCAATATAAA AAAGGTTCATAATTC |
| Length | 3615 |