CopciAB_505989
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_505989 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 505989 |
| Uniprot id | Functional description | Belongs to the glycosyl hydrolase 5 (cellulase A) family | |
| Location | scaffold_6:2069111..2071456 | Strand | + |
| Gene length (nt) | 2346 | Transcript length (nt) | 1499 |
| CDS length (nt) | 1419 | Protein length (aa) | 472 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | exgB |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_118685 | 61.7 | 3.746E-209 | 649 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_205540 | 61.7 | 7.976E-209 | 648 |
| Agrocybe aegerita | Agrae_CAA7265173 | 61.8 | 1.343E-206 | 642 |
| Grifola frondosa | Grifr_OBZ73724 | 62.3 | 2.174E-199 | 621 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_118440 | 59.2 | 1.517E-194 | 607 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1590136 | 59.1 | 4.79E-191 | 597 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1043147 | 59.7 | 9.848E-190 | 593 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB18518 | 61.7 | 1.474E-188 | 590 |
| Pleurotus ostreatus PC9 | PleosPC9_1_89806 | 57.7 | 5.633E-187 | 585 |
| Lentinula edodes NBRC 111202 | Lenedo1_342137 | 58.6 | 1.033E-181 | 570 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_7148 | 57.9 | 1.325E-178 | 561 |
| Schizophyllum commune H4-8 | Schco3_2620053 | 57.9 | 7.446E-178 | 559 |
| Auricularia subglabra | Aurde3_1_1230494 | 56.9 | 3.195E-164 | 520 |
| Lentinula edodes B17 | Lened_B_1_1_14229 | 56.2 | 6.9E-146 | 466 |
| Flammulina velutipes | Flave_chr06AA00970 | 55.1 | 3.434E-80 | 275 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_505989.T0 |
| Description | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 84 | 428 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 21 | 0.9808 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR001547 | Glycoside hydrolase, family 5 |
| IPR017853 | Glycoside hydrolase superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| K22277 |
EggNOG
| COG category | Description |
|---|---|
| G | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH5 | GH5_15 |
Transcription factor
| Group |
|---|
| No records |
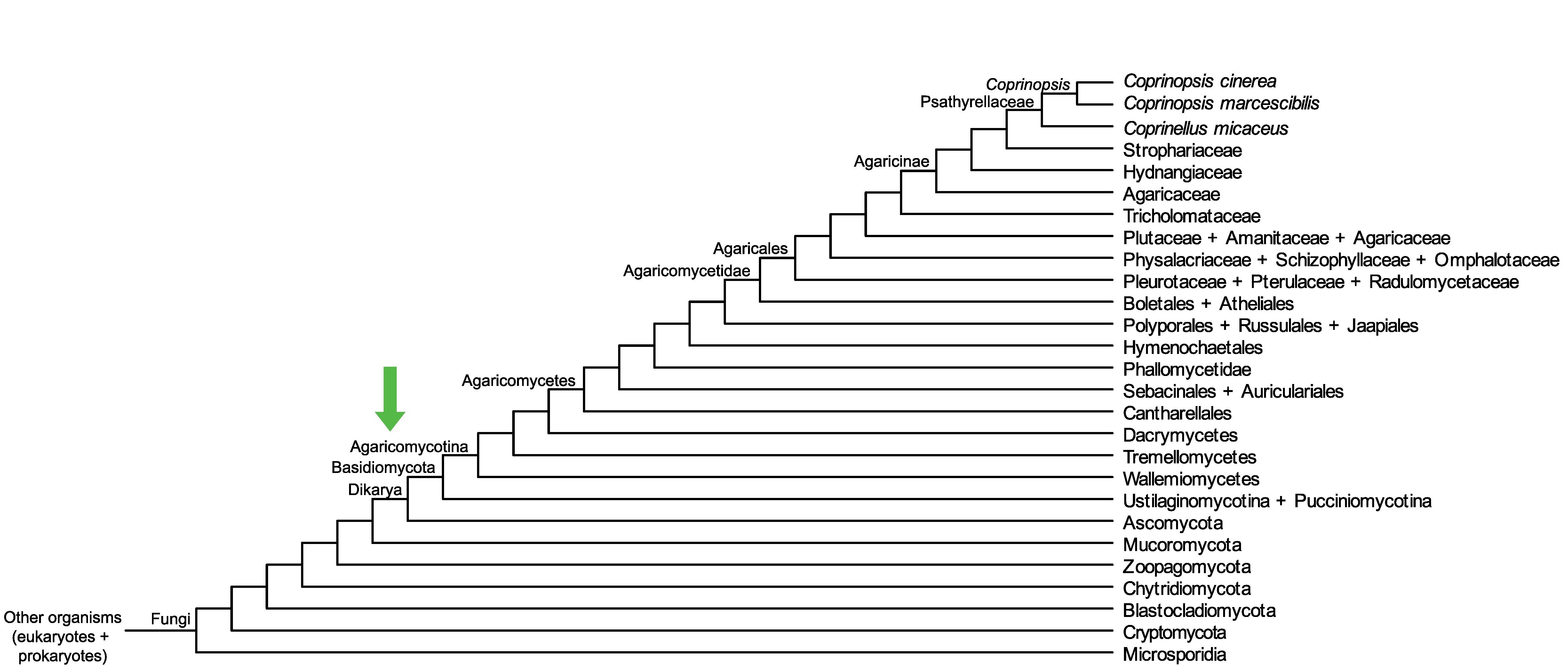
Conservation of CopciAB_505989 across fungi.
Arrow shows the origin of gene family containing CopciAB_505989.
Protein
| Sequence id | CopciAB_505989.T0 |
|---|---|
| Sequence |
>CopciAB_505989.T0 MPFTSFAHLVFLSALLAIVKATGLPSKIYGVNLGSWLVLEPWMLPQEWSNMGGQQCSDCSKCIASEFAFAQAYPT TVDAIFAKHWSTWFNQTDVNELVAAGINTVRIPLGYWIVEPLVDRRTEYYPKGGILHLKRGLRQLRDAGITVILD HHALPGVQSPNQMFAGRCTKDVQFYTPYNYRRALIWSGVITAISHLEPSFSSVASLQAINEPIMDANQTPFYGQF QKNFVQVVRAVELGLGIYIPGWHSANKVPPSTLSATDISTSLKSLSGATDLFNAEVRQVLREMIPVLFKVAAQLG VNTNFNKEGLRASLKRRSPLITNFMDMTWQYNNPSNPADAKIGPQTYDNHLYYSFGGVTDPNEEAYMTHICNLDR IEKAGALRNTPLWFGEWSLATQFGASDEFLHKWADAQKLAYSKSAGWIFWNFKIELSSRSGNTARQWSYMEGIRR GYFTKDPSKLNNPRVCDPYIRK |
| Length | 472 |
Coding
| Sequence id | CopciAB_505989.T0 |
|---|---|
| Sequence |
>CopciAB_505989.T0 ATGCCATTCACTTCGTTCGCCCATCTAGTTTTCTTGTCCGCTTTGCTGGCCATCGTCAAAGCTACTGGGCTGCCC AGCAAGATATATGGTGTTAACCTAGGATCTTGGTTGGTCTTGGAGCCATGGATGCTGCCACAGGAATGGTCCAAC ATGGGCGGGCAGCAATGTAGCGACTGCTCCAAGTGTATCGCATCAGAGTTCGCGTTTGCCCAGGCCTATCCCACA ACCGTCGACGCCATTTTTGCAAAACATTGGTCCACCTGGTTTAACCAAACCGACGTCAACGAACTGGTGGCTGCT GGAATCAACACCGTCCGAATTCCGCTTGGCTACTGGATTGTAGAGCCACTTGTCGATCGCAGAACTGAATATTAT CCTAAAGGCGGGATCCTGCACTTGAAAAGAGGCCTGAGACAACTTCGCGATGCAGGGATCACGGTCATCCTAGAC CACCACGCGCTGCCTGGCGTTCAATCGCCTAACCAAATGTTTGCAGGAAGGTGCACGAAAGACGTGCAGTTCTAT ACGCCATATAATTATCGCCGTGCGCTCATTTGGTCAGGGGTGATCACTGCAATCTCCCACCTTGAACCTTCCTTC AGCTCTGTAGCTTCATTGCAAGCTATTAACGAACCAATTATGGACGCGAACCAAACGCCATTCTACGGACAGTTC CAGAAGAATTTCGTTCAGGTCGTCCGCGCTGTCGAGTTGGGACTCGGAATCTACATCCCTGGTTGGCACAGTGCT AACAAAGTGCCCCCTTCCACTCTTTCTGCCACGGATATCAGTACCTCTTTGAAGTCGCTCTCTGGCGCAACCGAC CTTTTTAACGCGGAAGTAAGGCAGGTCCTTAGGGAGATGATTCCTGTACTTTTCAAGGTCGCTGCCCAACTCGGC GTGAACACCAACTTTAACAAAGAAGGATTAAGAGCTTCGTTGAAGAGGCGCTCGCCCTTAATCACCAACTTCATG GATATGACTTGGCAGTATAACAACCCTTCGAACCCTGCAGATGCGAAGATTGGCCCTCAGACATACGATAATCAT CTGTACTACTCTTTTGGGGGCGTCACTGACCCCAACGAAGAAGCGTACATGACTCACATATGCAACCTCGATCGA ATTGAGAAGGCGGGCGCTCTTAGGAACACACCTCTTTGGTTTGGAGAATGGAGTTTAGCCACCCAATTTGGCGCC TCGGATGAGTTCCTCCACAAATGGGCCGACGCCCAAAAGCTGGCGTATAGCAAGTCCGCTGGATGGATCTTCTGG AACTTCAAGATTGAGCTCTCATCACGATCTGGTAATACTGCGCGGCAGTGGTCCTATATGGAAGGCATCCGTCGG GGATATTTCACGAAAGACCCATCTAAACTGAACAATCCCAGGGTCTGCGATCCTTATATCAGGAAGTAA |
| Length | 1419 |
Transcript
| Sequence id | CopciAB_505989.T0 |
|---|---|
| Sequence |
>CopciAB_505989.T0 ACCACGATGCCATTCACTTCGTTCGCCCATCTAGTTTTCTTGTCCGCTTTGCTGGCCATCGTCAAAGCTACTGGG CTGCCCAGCAAGATATATGGTGTTAACCTAGGATCTTGGTTGGTCTTGGAGCCATGGATGCTGCCACAGGAATGG TCCAACATGGGCGGGCAGCAATGTAGCGACTGCTCCAAGTGTATCGCATCAGAGTTCGCGTTTGCCCAGGCCTAT CCCACAACCGTCGACGCCATTTTTGCAAAACATTGGTCCACCTGGTTTAACCAAACCGACGTCAACGAACTGGTG GCTGCTGGAATCAACACCGTCCGAATTCCGCTTGGCTACTGGATTGTAGAGCCACTTGTCGATCGCAGAACTGAA TATTATCCTAAAGGCGGGATCCTGCACTTGAAAAGAGGCCTGAGACAACTTCGCGATGCAGGGATCACGGTCATC CTAGACCACCACGCGCTGCCTGGCGTTCAATCGCCTAACCAAATGTTTGCAGGAAGGTGCACGAAAGACGTGCAG TTCTATACGCCATATAATTATCGCCGTGCGCTCATTTGGTCAGGGGTGATCACTGCAATCTCCCACCTTGAACCT TCCTTCAGCTCTGTAGCTTCATTGCAAGCTATTAACGAACCAATTATGGACGCGAACCAAACGCCATTCTACGGA CAGTTCCAGAAGAATTTCGTTCAGGTCGTCCGCGCTGTCGAGTTGGGACTCGGAATCTACATCCCTGGTTGGCAC AGTGCTAACAAAGTGCCCCCTTCCACTCTTTCTGCCACGGATATCAGTACCTCTTTGAAGTCGCTCTCTGGCGCA ACCGACCTTTTTAACGCGGAAGTAAGGCAGGTCCTTAGGGAGATGATTCCTGTACTTTTCAAGGTCGCTGCCCAA CTCGGCGTGAACACCAACTTTAACAAAGAAGGATTAAGAGCTTCGTTGAAGAGGCGCTCGCCCTTAATCACCAAC TTCATGGATATGACTTGGCAGTATAACAACCCTTCGAACCCTGCAGATGCGAAGATTGGCCCTCAGACATACGAT AATCATCTGTACTACTCTTTTGGGGGCGTCACTGACCCCAACGAAGAAGCGTACATGACTCACATATGCAACCTC GATCGAATTGAGAAGGCGGGCGCTCTTAGGAACACACCTCTTTGGTTTGGAGAATGGAGTTTAGCCACCCAATTT GGCGCCTCGGATGAGTTCCTCCACAAATGGGCCGACGCCCAAAAGCTGGCGTATAGCAAGTCCGCTGGATGGATC TTCTGGAACTTCAAGATTGAGCTCTCATCACGATCTGGTAATACTGCGCGGCAGTGGTCCTATATGGAAGGCATC CGTCGGGGATATTTCACGAAAGACCCATCTAAACTGAACAATCCCAGGGTCTGCGATCCTTATATCAGGAAGTAA TGGTTTGCGGCACCGATTGAACCACGGAGAGATAGCAATAATTCTCAGCTACAATGATTATCGAATTTAATACA |
| Length | 1499 |
Gene
| Sequence id | CopciAB_505989.T0 |
|---|---|
| Sequence |
>CopciAB_505989.T0 ACCACGATGCCATTCACTTCGTTCGCCCATCTAGTTTTCTTGTCCGCTTTGCTGGCCATCGTCAAAGCTACTGGG CTGCCCAGCAAGATATATGGTGTTAACCTAGGATCTTGGTGAGTCTTTCTAGTTATCTTTTTTGCGCGAAAATGA AGCCAACTTTTGTCCCAGGTTGGTCTTGGAGCCATGGATGCTGCCACAGGGTGTGTAGCGCCTGTTGGCCTCGTT CCATCACCCCGCCTAACATGATTTCCATCATACAGAATGGTCCAACATGGGCGGGCAGCAATGTAGCGACTGCTC CAAGTGTATCGCATCAGAGTTGTATGTCCACCATACCACCTTTCACCGCGGCTTCTTGACACTATGTGGGTAGCG CGTTTGCCCAGGCCTATCCCACAACCGTCGACGCCATTTTTGCAAAACATTGGTATGCGAAACCCAATATCCTGC CCAACAGTCCCAATGGTAATGAGAGCTAGGTCCACCTGGTTTAACCAAACCGACGTCAACGAACTGGTGGCTGCT GGAATCAACACCGTCCGAATTCCGGTAATGCTGCCTCAGGCTACCGTCATAGTATCAACCCCGAGACTAACCTAT TTATCCAGCTTGGCTACTGGATTGTAGAGCCACTTGTCGATCGCAGAACTGAATATTATCCTAAAGGCGGGATCC TGCACTTGGTGCGACTTTTTCTTGATTGATTAGTTGATTGTCAGAATTGACCTTCTTCGCACCTGCAGAAAAGAG GCCTGAGACAACTTCGCGATGCAGGGATCACGGTCATCCTAGACCACCACGCGCTGCCTGGCGTTCAATCGCCTA ACCAAATGTTTGCAGGAAGGTTAGCCTTCGCCTCATGGGTGGATGAATTGCTTCTGATGTCTATTTCATTTGTAT ACTAGGTGCACGAAAGACGTGCAGTTCTATGTACGTACACCTGCTCTTGCCTGCAGATGGTCGCTGAAGCCGCCA TCCATCAGACGCCATATAATTATCGCCGTGCGCTCATTTGGTCAGGGGTGATCACTGCAATCTCCCACCTTGAAC CTTCCTTCAGCTCTGTAGCTTCATTGCAAGCTATTAACGAACCAATTATGGACGCGAACCAAACGCCATTCTACG GACAGTGTAAGCTCCTGCACCACCCATGAACTTCGACTTGAACTGAATTTCGGCTAGTCCAGAAGAATTTCGTTC AGGTCGTCCGCGCTGTCGAGTTGGGACTCGGAATCTACATCCCTGGTTGGCACAGTGCTAACAAAGTGCCCCCTT CCACTCTTTCTGCCACGGATATCAGTACCTCTTTGAAGTCGCTCTCTGGCGCAACCGACCTTTTTAACGCGGAAG TAAGGCAGGTCCTTAGGGAGATGATTCCTGTACTTTTCAAGGTCGCTGCCCAACTCGGCGTGAACACCAACTTTA ACAAAGAAGGATTAAGAGCTTCGTTGAAGAGGCGCTCGCCCTTAATCACCAAGCAAGAACATCCTTGTTACCATA TTGCGCATCTACTGACTATCATATGTCGACCAGCTTCATGGATATGACTTGGCAGTATAACAACCCTTCGAACCC TGCAGATGCGAAGATTGGCCCTCAGACATACGATAATCATCTGTACTACTCTTTTGGGGTGGGTGTCTTGACGAC TGATTTGAACGTATCGGGCGAATAACGCGCCACACTTCACAGGGCGTCACTGACCCCAACGAAGAAGCGTACATG ACTCACATATGCAGTATGTTTTTCTATCTTTTTATCATGGGTGTACCATGGCTAATGGCTACCTTGACCAGACCT CGATCGAATTGAGAAGGCGGGCGCTCTTAGGAACACACCTCTTTGGTTTGGAGGTGAATTCCGCCCGAAATGACG GATTAATGCAATTGTCTAACGGACCTTGTCCCAGAATGGAGTTTAGCCACCCAATTTGGCGCCTCGGATGAGTTC CTCCACAAATGGGCCGACGCCCAAAAGCTGGCGTATAGCAAGTCCGCTGGATGGATCGTAAGTTGTTTGGTTTGA ACTTTGCAGACTGTTGACGTATTGTCCATCCAGTTCTGGAACTTCAAGATTGAGCTCTCATCACGATCTGGTAAT ACTGCGCGGCAGTGGTGAGTGCTTGTACCCTCTCAGGGTTATCTTAAGTTCCTGATGTACTTTGGCGCGCTTCGC TAGGTCCTATATGGAAGGCATCCGTCGGGGATATTTCACGAAAGACCCATCTAAACTGAACAATCCCAGGGTCTG CGATCCTTATATCAGGAAGTAATGGTTTGCGGCACCGATTGAACCACGGAGAGATAGCAATAATTCTCAGCTACA ATGATTATCGAATTTAATACA |
| Length | 2346 |