CopciAB_545273
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_545273 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | rgxA | Synonyms | 545273 |
| Uniprot id | Functional description | Belongs to the glycosyl hydrolase 28 family | |
| Location | scaffold_7:363090..365761 | Strand | + |
| Gene length (nt) | 2672 | Transcript length (nt) | 1668 |
| CDS length (nt) | 1302 | Protein length (aa) | 433 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_194940 | 57.9 | 9.554E-160 | 504 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_121554 | 57.6 | 2.207E-159 | 503 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB28139 | 56.1 | 3.367E-159 | 503 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_146110 | 54.2 | 8.19E-155 | 490 |
| Pleurotus eryngii ATCC 90797 | Pleery1_451035 | 50.3 | 1.373E-149 | 475 |
| Grifola frondosa | Grifr_OBZ72754 | 54.6 | 1.359E-148 | 472 |
| Agrocybe aegerita | Agrae_CAA7268813 | 53.3 | 1.832E-146 | 466 |
| Lentinula edodes NBRC 111202 | Lenedo1_1033606 | 49.8 | 2.142E-134 | 431 |
| Auricularia subglabra | Aurde3_1_146384 | 50.1 | 9.043E-132 | 424 |
| Pleurotus ostreatus PC9 | PleosPC9_1_82769 | 46.7 | 5.975E-129 | 415 |
| Schizophyllum commune H4-8 | Schco3_235495 | 49.1 | 1.94E-127 | 411 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1020172 | 47.7 | 3.512E-126 | 407 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_22344 | 55.5 | 3.141E-109 | 358 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | rgxA |
|---|---|
| Protein id | CopciAB_545273.T0 |
| Description | Belongs to the glycosyl hydrolase 28 family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00295 | Glycosyl hydrolases family 28 | IPR000743 | 103 | 422 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 22 | 0.9992 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011050 | Pectin lyase fold/virulence factor |
| IPR000743 | Glycoside hydrolase, family 28 |
| IPR012334 | Pectin lyase fold |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004650 | polygalacturonase activity | MF |
| GO:0005975 | carbohydrate metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| G | Belongs to the glycosyl hydrolase 28 family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH28 |
Transcription factor
| Group |
|---|
| No records |
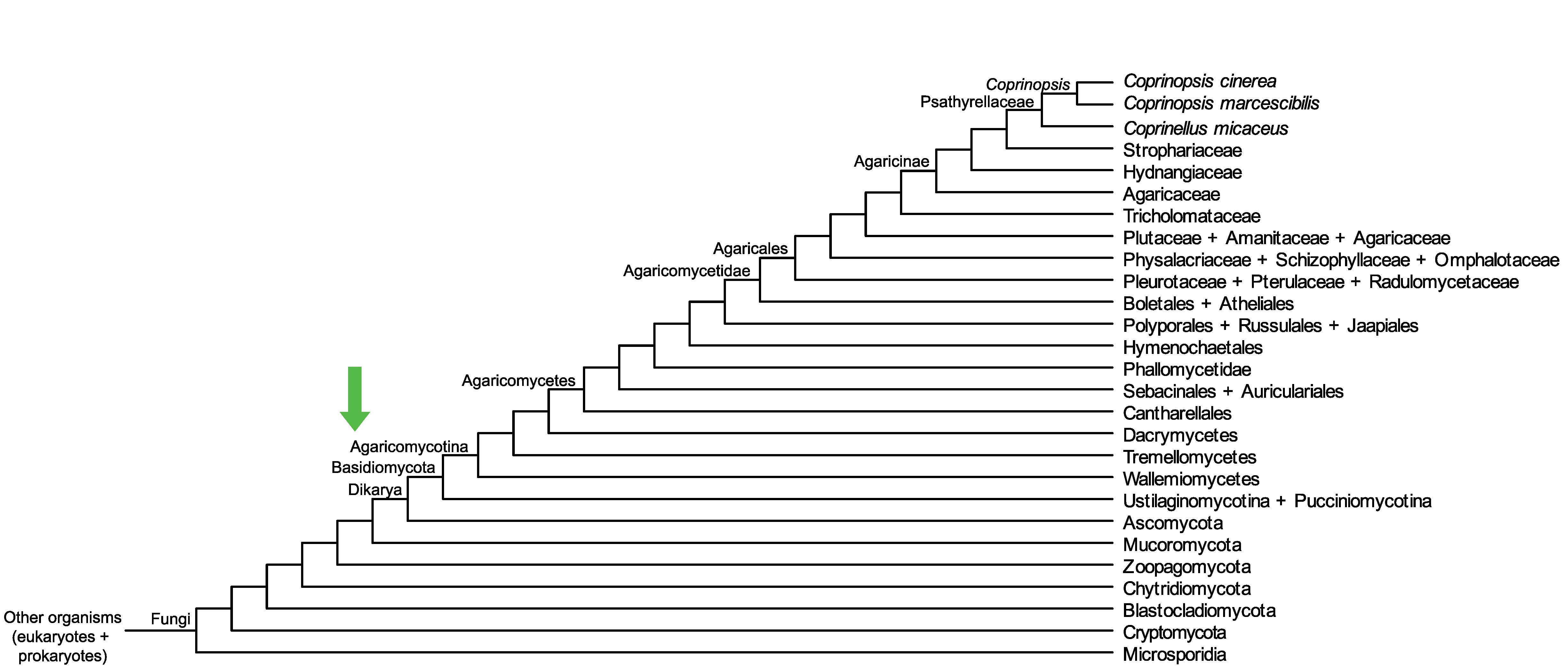
Conservation of CopciAB_545273 across fungi.
Arrow shows the origin of gene family containing CopciAB_545273.
Protein
| Sequence id | CopciAB_545273.T0 |
|---|---|
| Sequence |
>CopciAB_545273.T0 MVLKSSWLLALCSLAVLGPVSGTPVDLDVATDKGKVCRLKPLGRGRDDTDQVEAAIKRCGNGGTTIFEEGDYKIT RKLFWELNSARVELKGRLSFEPKVEYWLDPANTFRVVFIQSQASWFVVTGKNFVIDGKKKGGIEGNGQPWWEYFT VVPRQDGNGRPIAFTLWKAKNAVIKDFNIISPPFWANAVAESSEILYDGMFVNATNLNPKYFGQNVVWNTDGIDT YRSERITMKNWDVTSGDDCIAVKGNSSKIVARDFVCRGGTGIAFGSLGQYAHLTDYVTDVLVENFKVLRLPSRQQ PNMNYGIYFKAWDGPITGTPPTGGGGGRGLVSNAIFRNVIFDRVMTGVQIWQNFGSDGEAPVTSKVKFDGLLFEN IKGTTNTSNIVRLECSSEAPCSNFTFRNFRVHGPNKTESTYICDNTENIRGLPAPCNN |
| Length | 433 |
Coding
| Sequence id | CopciAB_545273.T0 |
|---|---|
| Sequence |
>CopciAB_545273.T0 ATGGTCCTCAAATCCAGTTGGCTTTTGGCGCTCTGTAGTCTCGCTGTTCTTGGACCTGTCTCTGGGACGCCTGTT GATCTTGATGTCGCCACCGACAAAGGAAAAGTGTGTCGATTGAAACCGTTGGGACGAGGAAGAGATGACACCGAT CAGGTCGAAGCTGCGATTAAGCGCTGTGGAAATGGCGGTACAACCATCTTTGAAGAGGGAGACTACAAAATTACA AGGAAACTCTTCTGGGAACTCAACTCGGCTCGAGTAGAGCTCAAAGGACGATTAAGCTTCGAGCCGAAGGTGGAG TACTGGCTCGACCCAGCCAACACCTTCCGGGTGGTCTTCATCCAAAGCCAAGCCTCCTGGTTCGTCGTAACCGGC AAGAACTTTGTGATCGACGGAAAGAAGAAGGGTGGGATTGAGGGGAACGGCCAGCCTTGGTGGGAATACTTTACG GTAGTCCCAAGGCAGGACGGGAACGGGCGGCCCATCGCGTTCACGCTCTGGAAAGCGAAGAATGCTGTCATCAAG GATTTCAACATCATTTCGCCACCGTTCTGGGCTAATGCGGTGGCCGAATCGAGTGAGATTCTGTATGATGGCATG TTCGTCAATGCTACGAACTTGAATCCCAAGTACTTTGGACAAAATGTTGTCTGGAATACCGATGGAATTGATACA TATCGAAGCGAACGTATCACGATGAAGAATTGGGATGTTACCAGCGGAGATGATTGCATTGCCGTCAAGGGCAAT TCTTCCAAGATCGTTGCCCGAGACTTTGTTTGTCGGGGCGGAACTGGGATCGCATTCGGCTCGCTAGGCCAATAC GCTCACTTGACGGATTATGTCACCGATGTCTTGGTCGAGAATTTCAAGGTGTTGCGCCTTCCTTCAAGGCAACAA CCAAACATGAACTACGGTATCTACTTCAAAGCTTGGGACGGTCCAATCACCGGTACTCCTCCAACTGGAGGAGGC GGAGGACGAGGACTCGTCAGCAACGCCATTTTCCGAAACGTCATCTTTGACCGGGTCATGACTGGCGTCCAGATC TGGCAAAACTTTGGTAGCGACGGAGAAGCGCCCGTAACCAGCAAAGTGAAGTTTGACGGCCTCCTTTTTGAGAAC ATAAAGGGAACTACGAACACCAGTAACATCGTACGATTGGAGTGCAGCTCTGAAGCCCCTTGCTCCAACTTCACC TTCCGCAACTTCCGAGTCCACGGCCCCAACAAGACTGAATCGACCTACATCTGTGACAACACCGAAAACATTCGA GGACTACCAGCACCTTGCAACAATTAA |
| Length | 1302 |
Transcript
| Sequence id | CopciAB_545273.T0 |
|---|---|
| Sequence |
>CopciAB_545273.T0 CTCATGACCAAGGCCTTTTCCTTCGAACTTCTCTAAGTCTGGCTCGACCATGACATCTCTTCACATTCGAACTGA AAATGCCCCCAAATCGCAGAAATAGATGGCCCTTCCTCGGCATATCTACCTCGCTCGAGTAGCTATTAACCTTGG ACTGTCGACGTTACTTGTTCGAGATACTTGTCTTTGTTTTTTGTCTCTGGAACCCTACACACCTCTCGAGATACT CTACAATGGTCCTCAAATCCAGTTGGCTTTTGGCGCTCTGTAGTCTCGCTGTTCTTGGACCTGTCTCTGGGACGC CTGTTGATCTTGATGTCGCCACCGACAAAGGAAAAGTGTGTCGATTGAAACCGTTGGGACGAGGAAGAGATGACA CCGATCAGGTCGAAGCTGCGATTAAGCGCTGTGGAAATGGCGGTACAACCATCTTTGAAGAGGGAGACTACAAAA TTACAAGGAAACTCTTCTGGGAACTCAACTCGGCTCGAGTAGAGCTCAAAGGACGATTAAGCTTCGAGCCGAAGG TGGAGTACTGGCTCGACCCAGCCAACACCTTCCGGGTGGTCTTCATCCAAAGCCAAGCCTCCTGGTTCGTCGTAA CCGGCAAGAACTTTGTGATCGACGGAAAGAAGAAGGGTGGGATTGAGGGGAACGGCCAGCCTTGGTGGGAATACT TTACGGTAGTCCCAAGGCAGGACGGGAACGGGCGGCCCATCGCGTTCACGCTCTGGAAAGCGAAGAATGCTGTCA TCAAGGATTTCAACATCATTTCGCCACCGTTCTGGGCTAATGCGGTGGCCGAATCGAGTGAGATTCTGTATGATG GCATGTTCGTCAATGCTACGAACTTGAATCCCAAGTACTTTGGACAAAATGTTGTCTGGAATACCGATGGAATTG ATACATATCGAAGCGAACGTATCACGATGAAGAATTGGGATGTTACCAGCGGAGATGATTGCATTGCCGTCAAGG GCAATTCTTCCAAGATCGTTGCCCGAGACTTTGTTTGTCGGGGCGGAACTGGGATCGCATTCGGCTCGCTAGGCC AATACGCTCACTTGACGGATTATGTCACCGATGTCTTGGTCGAGAATTTCAAGGTGTTGCGCCTTCCTTCAAGGC AACAACCAAACATGAACTACGGTATCTACTTCAAAGCTTGGGACGGTCCAATCACCGGTACTCCTCCAACTGGAG GAGGCGGAGGACGAGGACTCGTCAGCAACGCCATTTTCCGAAACGTCATCTTTGACCGGGTCATGACTGGCGTCC AGATCTGGCAAAACTTTGGTAGCGACGGAGAAGCGCCCGTAACCAGCAAAGTGAAGTTTGACGGCCTCCTTTTTG AGAACATAAAGGGAACTACGAACACCAGTAACATCGTACGATTGGAGTGCAGCTCTGAAGCCCCTTGCTCCAACT TCACCTTCCGCAACTTCCGAGTCCACGGCCCCAACAAGACTGAATCGACCTACATCTGTGACAACACCGAAAACA TTCGAGGACTACCAGCACCTTGCAACAATTAAGGGTTGAGTCCCTTCGAGTCTCATCAGGAGTAAAGCCTGAAAA GGAGCCTCTTTTGCGTAATGAACTACCTATTGTGTACCTGCCTACAGCCTCTATCGAAAATACCGTGAATAAACG CTGAATTTAGCGCCCTAA |
| Length | 1668 |
Gene
| Sequence id | CopciAB_545273.T0 |
|---|---|
| Sequence |
>CopciAB_545273.T0 CTCATGACCAAGGCCTTTTCCTTCGAACTTCTCTAAGTCTGGCTCGACCATGACATCTCTTCACATTCGAACTGA AAATGCCCCCAAATCGCAGAAATAGATGGCCCTTCCTCGGCATATCTACCTCGCTCGAGTAGCTATTAACCTTGG ACTGTCGACGTTACTTGTTCGAGATACTTGTCTTTGTTTTTTGTCTCTGGAACCCTACACACCTCTCGAGATACT CTACAATGGTCCTCAAATCCAGTTGGCTTTTGGCGCTCTGTAGTCTCGCTGTTCTTGGACCTGTCTCTGGGACGC CTGTTGATCTTGATGTCGCCACCGACAAAGGAAAAGTGTGTCGATTGAAACCGTTGGGACGAGGAAGAGATGACA CCGATCAGGTTCGTTTAAACTAGGTCACTATCTAGCAGTTGTACGCCGGATTGACATGACTTGCTAGGTCGAAGC TGCGATTAAGCGCTGTGGAAATGGCGGTACAACCATCTTTGAAGAGGGAGACTACAAAATTACAAGGTCAGCGAC TACCTCGGGGTGTCAAGATTGCCGCTGAGCTACAAAAACATCTTCCTCTAGGAAACTCTTCTGGGAACTCAACTC GGCTCGAGTAGAGCTCAAAGGACGATTAAGCGTGAGCCTTTCTATCCATCCACAACCCTTCCTACTCTTCTTCTT ACTCTTCGAACATCTTCTGATCCCTCTTCTCATCTAGTTCGAGCCGAAGGTGGAGTACTGGCTCGACCCAGCCAA CACCTTCCGGGTGGTCTTCATCCAAGTACGTACATATTTCTGCGTACTGTATATCTACAACCGTCCCGCCCCGCA CCGGTGCTGTCCGGCTAACCCGTCATGATCATCACATCGCAGAGCCAAGCCTCCTGGTTCGTCGTAACCGGCAAG AACTTTGTGATCGACGGAAAGAAGAAGGGTGGGATTGAGGGGAACGGCCAGGCAAGTCCACCTACAACCTTCAAT TTCCTTCGCACCTTCGAAGCATGGAGAATTGATTTTCCCGATTCCAATTCCGATTCTATTATCACTTTCTGAATC TGTGCCAATCATGAATCATGGCCCGGGCAGCCTTGGTGGGAATACTTTACGGTAGTCCCAAGGCAGGACGGGAAC GGGCGGCCCATCGCGTTCACGCTCTGGAAAGCGAAGAATGCTGTCATCAAGGATTTCAACATCATTTCGCCACCG TTCTGGGCTAATGCGGTGGCCGAATCGAGTGAGATTCTGTATGATGGCATGTTCGTCAATGCTACGAACTTGAAT CCCAAGTACTTTGGACAAAAGTGAGAGCTTCAAGTATCGTTCTAACTTCGGGGGGCTATGCGATTGACTTTTTTT TCTTGACAGTGTTGTCTGGAATACCGATGGTCAGCAGCGATAAACTCCATGTTGTTGATGTTTGGCTTACATCCA ACAGGAATTGATACATATCGAAGCGAACGTATCACGATGAAGAATTGGGATGTTACCAGCGGAGATGATTGCATT GCCGTCAAGGGCGTAAGTTTAGACCAGACTCCAGAGACCATCTACCTAGCTGCATGCTGACTCTGTAATTGCAGA ATTCTTCCAAGATCGTTGCCCGAGACTTTGTTTGTCGGGGCGGAACTGGGATCGCATTCGGCTCGCTAGGCCAAT ACGCTCACTTGGTAGGGTTCCTGTTCTACATTTATCTTCTTATCCTCTGTGCTAATACAGGGCGCAGACGGATTA TGTCACCGATGTCTTGGTCGAGAATTTCAAGGTCGGTATCATCGCTGCCACCTGCTACCCCCAATACATTCTCAT TCTCGACGAACACAGGTGTTGCGCCTTCCTTCAAGGCAACAACCAAACATGAACTACGGTGTAGGCCATGCTCAA AATTAGGATTACTTCATCTATACTCACAACGCTCTAGATCTACTTCAAAGCTTGGGACGGTCCAATCACCGGTAC TCCTCCAACTGGAGGAGGCGGAGGACGAGGTAATCCAAATCCCTCTCCACAATCACAAACCTTCTCTCTAAACTG CAAACTCCGTTATAGGACTCGTCAGCAACGCCATTTTCCGAAACGTCATCTTTGACCGGGTCATGACTGGCGTCC AGATCTGGCAAAACTTTGGTAGCGAGTACGTTCCAAAGGATATTTCCCCCTCCACCGTCCAAACCTAACATGACC GTTACTTTCCTCAGCGGAGAAGCGCCCGTAACCAGCAAAGTGAAGTTTGACGGCCTCCTTTTTGAGAACATAAAG GGAACTACGAACACCAGTAACAGTACGTCGAACCCACTCGAGCGTTGACGGGACCAAGTTGACCCCAGTGGCTCT ACTACGACAGTCGTACGATTGGAGTGCAGCTCTGAAGCCCCTTGCTCCAACTTCACCTTCCGCAACTTCCGAGTC CACGGCCCCAACAAGACTGAATCGACCTACATCTGTGACAACACCGAAAACATTCGAGGACTACCAGGTCAGTTT CATTTGCGCCACACTTACCTCGAAGGTTCTGACGGTCGTTTTAGCACCTTGCAACAATTAAGGGTTGAGTCCCTT CGAGTCTCATCAGGAGTAAAGCCTGAAAAGGAGCCTCTTTTGCGTAATGAACTACCTATTGTGTACCTGCCTACA GCCTCTATCGAAAATACCGTGAATAAACGCTGAATTTAGCGCCCTAA |
| Length | 2672 |