CopciAB_546300
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_546300 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | EXG3 | Synonyms | 546300 |
| Uniprot id | Functional description | Belongs to the glycosyl hydrolase 5 (cellulase A) family | |
| Location | scaffold_2:2724218..2726516 | Strand | + |
| Gene length (nt) | 2299 | Transcript length (nt) | 2112 |
| CDS length (nt) | 1749 | Protein length (aa) | 582 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Schizosaccharomyces pombe | exg3 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB24049 | 48.1 | 2.029E-182 | 578 |
| Agrocybe aegerita | Agrae_CAA7265374 | 46.9 | 5.023E-169 | 539 |
| Schizophyllum commune H4-8 | Schco3_2613657 | 43 | 4.565E-161 | 516 |
| Grifola frondosa | Grifr_OBZ75975 | 43.6 | 1.6E-151 | 488 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_200333 | 45.6 | 3.069E-150 | 484 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1381013 | 44.5 | 2.192E-148 | 479 |
| Lentinula edodes NBRC 111202 | Lenedo1_1094061 | 43.3 | 1.384E-132 | 433 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_8028 | 42 | 3.465E-130 | 426 |
| Pleurotus ostreatus PC9 | PleosPC9_1_82312 | 42.9 | 1.975E-124 | 409 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1033087 | 50.4 | 2.152E-123 | 406 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_53834 | 48.2 | 2.044E-94 | 321 |
| Lentinula edodes B17 | Lened_B_1_1_297 | 35.9 | 5.69E-89 | 305 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | EXG3 |
|---|---|
| Protein id | CopciAB_546300.T0 |
| Description | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 151 | 426 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 19 | 0.9039 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR017853 | Glycoside hydrolase superfamily |
| IPR001547 | Glycoside hydrolase, family 5 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| K01210 |
EggNOG
| COG category | Description |
|---|---|
| G | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH5 | GH5_50 |
Transcription factor
| Group |
|---|
| No records |
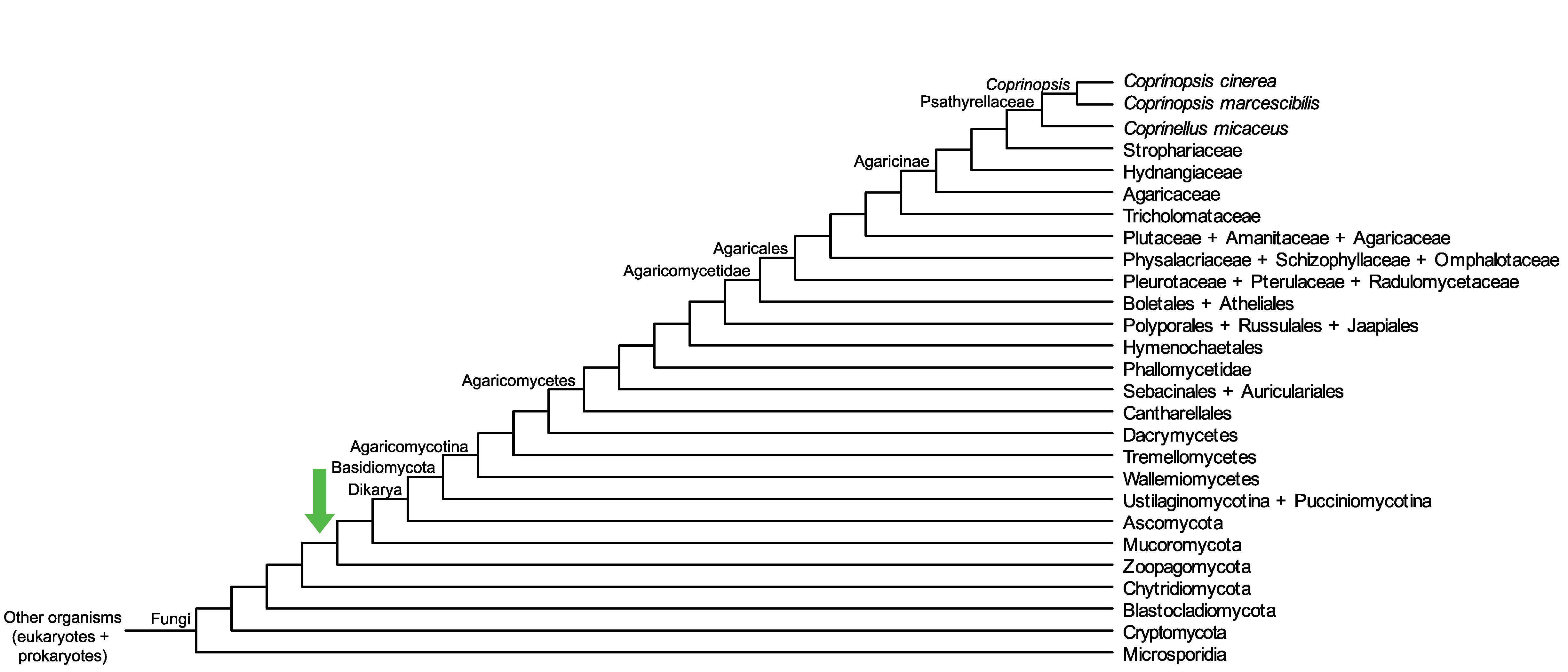
Conservation of CopciAB_546300 across fungi.
Arrow shows the origin of gene family containing CopciAB_546300.
Protein
| Sequence id | CopciAB_546300.T0 |
|---|---|
| Sequence |
>CopciAB_546300.T0 MVSNATFVSALLALGVTQANYKPYPSVSNTSTSTTRTISTVDDTDVQPDGLVLPQNVRITAVMDASDPQCQVEPY DAPSVDHQEYPSYDEKLATVFRYRKQQSVNLGSWFVHENWMTPSLFRCAAGNAGSEIDIAYGWGNTTGARTVLER HWDTFINASDFKWLADVGINTVRLPIGYWNLGSDFVKGTDYESAAEVYQNSWARVKRAVNLAGEHGLGVLVDLHG VPGSQNGKDHSGVSNGASNLFGDSANMDKTIDILTFLTKEFVHVNNVVGIQVLNEPIFDDRLTDFYGRAMDAMRA ADPDASRLPLYAHNGFDLKRFGPFVTGRKDFVVQDHHSYFVFSPEDRDQTATDHANSISNDVASTLGNASQETRG ELIVGEWSCALPPESLASDSNQNQAHKDFCGGQVDTYGNNTAGWSFWSYTKEECDNDPGWCFKSAVGTVLPTKFF SYHHQQPESIQNSVAGSNHSFLLHDAELPLTRRDEFTRLSIPERFKTIARRRRRDGNTDADGKGYDDGRATAGIF AAHNMSRLGFVNQYISDTIANLGSNVIPQESEGAYTEGFLRGLREAESNFSTQAFTS |
| Length | 582 |
Coding
| Sequence id | CopciAB_546300.T0 |
|---|---|
| Sequence |
>CopciAB_546300.T0 ATGGTCTCCAACGCTACTTTTGTTTCTGCTTTACTGGCGCTGGGCGTTACTCAAGCAAACTACAAACCATACCCC AGCGTTTCGAACACCTCGACATCAACAACAAGAACAATATCCACTGTCGACGACACCGACGTACAACCAGATGGC CTCGTTTTGCCTCAGAATGTCCGCATTACAGCCGTCATGGATGCGTCGGATCCCCAGTGTCAGGTCGAACCCTAT GATGCACCCTCAGTGGACCATCAAGAATACCCCTCATATGACGAGAAACTCGCTACAGTGTTTCGATATCGGAAA CAACAGTCTGTGAATCTAGGGTCTTGGTTTGTTCACGAGAATTGGATGACCCCATCGCTCTTCCGCTGTGCGGCA GGCAATGCTGGCTCAGAAATCGACATCGCCTATGGGTGGGGAAACACAACTGGCGCTCGTACTGTTTTGGAACGA CATTGGGATACCTTCATCAACGCTTCGGACTTCAAATGGTTGGCAGATGTGGGTATCAATACCGTTCGCCTGCCC ATCGGATATTGGAATCTAGGGTCAGATTTCGTCAAAGGGACGGATTACGAGAGTGCAGCCGAAGTGTATCAGAAC TCTTGGGCAAGGGTGAAGAGGGCGGTGAACCTGGCTGGAGAACATGGGCTTGGTGTCTTGGTCGACCTGCATGGC GTTCCCGGAAGCCAGAACGGAAAGGACCACTCGGGGGTCTCGAATGGTGCTTCCAACCTCTTCGGCGACTCCGCG AACATGGACAAGACCATCGATATCCTTACGTTCCTAACGAAGGAGTTTGTTCATGTGAACAATGTTGTCGGCATT CAAGTCCTTAACGAACCAATATTCGATGACCGTTTGACTGATTTCTATGGCAGAGCCATGGACGCTATGCGGGCG GCTGATCCTGACGCCTCTCGCCTTCCGTTATATGCCCATAATGGGTTCGACTTGAAACGTTTTGGCCCATTTGTT ACTGGACGGAAGGATTTTGTCGTGCAGGATCATCATTCGTACTTTGTCTTCAGCCCAGAGGACAGGGACCAAACC GCGACAGACCATGCGAACAGTATCTCCAATGATGTCGCGTCGACGTTGGGTAACGCTTCGCAAGAAACCAGGGGG GAGCTCATTGTCGGCGAATGGTCCTGCGCCCTTCCACCAGAAAGCCTTGCATCCGATTCGAATCAAAATCAAGCG CATAAAGATTTCTGCGGCGGGCAGGTTGATACCTATGGAAACAACACTGCTGGTTGGAGCTTCTGGTCTTACACG AAAGAAGAATGCGATAACGATCCCGGGTGGTGCTTCAAGTCAGCGGTCGGCACTGTTTTACCCACGAAGTTCTTC TCGTACCATCATCAACAACCAGAGTCCATCCAAAACAGCGTTGCTGGATCCAATCATTCCTTCCTGCTTCATGAC GCGGAGCTGCCGTTAACCAGGCGTGACGAGTTTACGAGGCTCTCTATTCCCGAGCGCTTCAAAACCATCGCTAGA AGACGCAGGCGAGATGGAAACACAGATGCAGATGGTAAAGGGTATGACGACGGCAGGGCGACGGCTGGCATATTT GCTGCGCATAACATGTCAAGGCTGGGATTCGTCAACCAATACATCTCGGATACCATCGCCAACCTTGGATCCAAT GTTATACCCCAGGAATCTGAAGGTGCTTATACAGAGGGATTCCTTCGTGGTCTTCGGGAGGCAGAGTCGAATTTC TCGACACAAGCTTTCACATCGTAG |
| Length | 1749 |
Transcript
| Sequence id | CopciAB_546300.T0 |
|---|---|
| Sequence |
>CopciAB_546300.T0 GACGTTCTCTTCTGCCTCCACTCGGCTGTGCCCTTGTCCCGTTCATATCTCTGTATTTTTTCAAGGCCATTATTC AGTCATGGATGGCCTATGGTTGAAGCTTGTAGCACTCAATGGGCGACCGGGTCCTGAGTCATCCGGAATATCGAT CTCGACGATTGCTTAGCTCCTCTCGATCACCCCTATATAACCAGAGGAACTTGTCCCCGGATGGGCACCTTTTCA CCAAAGTCTACACGCTTTTGAAGAATGGTCTCCAACGCTACTTTTGTTTCTGCTTTACTGGCGCTGGGCGTTACT CAAGCAAACTACAAACCATACCCCAGCGTTTCGAACACCTCGACATCAACAACAAGAACAATATCCACTGTCGAC GACACCGACGTACAACCAGATGGCCTCGTTTTGCCTCAGAATGTCCGCATTACAGCCGTCATGGATGCGTCGGAT CCCCAGTGTCAGGTCGAACCCTATGATGCACCCTCAGTGGACCATCAAGAATACCCCTCATATGACGAGAAACTC GCTACAGTGTTTCGATATCGGAAACAACAGTCTGTGAATCTAGGGTCTTGGTTTGTTCACGAGAATTGGATGACC CCATCGCTCTTCCGCTGTGCGGCAGGCAATGCTGGCTCAGAAATCGACATCGCCTATGGGTGGGGAAACACAACT GGCGCTCGTACTGTTTTGGAACGACATTGGGATACCTTCATCAACGCTTCGGACTTCAAATGGTTGGCAGATGTG GGTATCAATACCGTTCGCCTGCCCATCGGATATTGGAATCTAGGGTCAGATTTCGTCAAAGGGACGGATTACGAG AGTGCAGCCGAAGTGTATCAGAACTCTTGGGCAAGGGTGAAGAGGGCGGTGAACCTGGCTGGAGAACATGGGCTT GGTGTCTTGGTCGACCTGCATGGCGTTCCCGGAAGCCAGAACGGAAAGGACCACTCGGGGGTCTCGAATGGTGCT TCCAACCTCTTCGGCGACTCCGCGAACATGGACAAGACCATCGATATCCTTACGTTCCTAACGAAGGAGTTTGTT CATGTGAACAATGTTGTCGGCATTCAAGTCCTTAACGAACCAATATTCGATGACCGTTTGACTGATTTCTATGGC AGAGCCATGGACGCTATGCGGGCGGCTGATCCTGACGCCTCTCGCCTTCCGTTATATGCCCATAATGGGTTCGAC TTGAAACGTTTTGGCCCATTTGTTACTGGACGGAAGGATTTTGTCGTGCAGGATCATCATTCGTACTTTGTCTTC AGCCCAGAGGACAGGGACCAAACCGCGACAGACCATGCGAACAGTATCTCCAATGATGTCGCGTCGACGTTGGGT AACGCTTCGCAAGAAACCAGGGGGGAGCTCATTGTCGGCGAATGGTCCTGCGCCCTTCCACCAGAAAGCCTTGCA TCCGATTCGAATCAAAATCAAGCGCATAAAGATTTCTGCGGCGGGCAGGTTGATACCTATGGAAACAACACTGCT GGTTGGAGCTTCTGGTCTTACACGAAAGAAGAATGCGATAACGATCCCGGGTGGTGCTTCAAGTCAGCGGTCGGC ACTGTTTTACCCACGAAGTTCTTCTCGTACCATCATCAACAACCAGAGTCCATCCAAAACAGCGTTGCTGGATCC AATCATTCCTTCCTGCTTCATGACGCGGAGCTGCCGTTAACCAGGCGTGACGAGTTTACGAGGCTCTCTATTCCC GAGCGCTTCAAAACCATCGCTAGAAGACGCAGGCGAGATGGAAACACAGATGCAGATGGTAAAGGGTATGACGAC GGCAGGGCGACGGCTGGCATATTTGCTGCGCATAACATGTCAAGGCTGGGATTCGTCAACCAATACATCTCGGAT ACCATCGCCAACCTTGGATCCAATGTTATACCCCAGGAATCTGAAGGTGCTTATACAGAGGGATTCCTTCGTGGT CTTCGGGAGGCAGAGTCGAATTTCTCGACACAAGCTTTCACATCGTAGTGCTTTACCAACCTTGCATTCGCTATT CGATTTTCACCGTTGTATTGCTAGCACCATTTATTTTACTCTGGGAGTACTTTGCTCGCCCGGTGTTCACAACCA GACATTTCAGTA |
| Length | 2112 |
Gene
| Sequence id | CopciAB_546300.T0 |
|---|---|
| Sequence |
>CopciAB_546300.T0 GACGTTCTCTTCTGCCTCCACTCGGCTGTGCCCTTGTCCCGTTCATATCTCTGTATTTTTTCAAGGCCATTATTC AGTCATGGATGGCCTATGGTTGAAGCTTGTAGCACTCAATGGGCGACCGGGTCCTGAGTCATCCGGAATATCGAT CTCGACGATTGCTTAGCTCCTCTCGATCACCCCTATATAACCAGAGGAACTTGTCCCCGGATGGGCACCTTTTCA CCAAAGTCTACACGCTTTTGAAGAATGGTCTCCAACGCTACTTTTGTTTCTGCTTTACTGGCGCTGGGCGTTACT CAAGCAAACTACAAACCATACCCCAGCGTTTCGAACACCTCGACATCAACAACAAGAACAATATCCACTGTCGAC GACACCGACGTACAACCAGATGGCCTCGTTTTGCCTCAGAATGTCCGCATTACAGCCGTCATGGATGCGTCGGAT CCCCAGTGTCAGGTCGAACCCTATGATGCACCCTCAGTGGACCATCAAGAATACCCCTCATATGACGAGAAACTC GCTACAGTGTTTCGATATCGGAAACAACAGTCTGTGAATCTAGGGTCTTGGTACGTGGTCAATTTGCCTATCCGC CGTGACGTCTCTTTAGAACCCTTTCCTAACCCTTGCCCCGCAATAGGTTTGTTCACGAGAATTGGATGACCCCAT CGCTCTTCCGCTGTGCGGCAGGCAATGCTGGCTCAGAAATCGACATCGCCTATGGGTGGGGAAACACAACTGGCG CTCGTACTGTTTTGGAACGACATTGGGATACCTTCATCAACGCTTCGGACTTCAAATGGTTGGCAGATGTGGGTA TCAATACCGTTCGCCTGCCCATCGGATATTGGAATCTAGGGTCAGATTTCGTCAAAGGGACGGATTACGAGAGTG CAGCCGAAGTGTATCAGAACTCTTGGGCAAGGGTGAAGAGGGCGGTGAACCTGGCTGGAGAACATGGGCTTGGTG TCTTGGTCGACCTGCATGGCGTTCCCGGAAGCCAGAACGGAAAGGACCACTCGGGGGTCTCGAATGGTGCTTCCA ACCTCTTCGGCGACTCCGCGAACATGGACAAGACCATCGATATCCTTACGTTCCTAACGAAGGAGTTTGTTCATG TGAACAATGTTGTCGGCATTCAAGTCCTTAACGAACCAATATTCGATGACCGTTTGACTGATTTCTGTGAGTATT TCTTCCTTGAGACGTTGACATCAACTTATAGACATCTTGTAGATGGCAGAGCCATGGACGCTATGCGGGCGGCTG ATCCTGACGCCTCTCGCCTTCCGTTATATGCCCATAATGGGTTCGACTTGAAACGTTTTGGCCCATTTGTTACTG GACGGAAGGATTTTGTCGTGCAGGATCATCATTCGTACTTTGTCTTCAGCCCAGAGGACAGGGACCAAACCGCGA CAGACCATGCGAACAGTATCTCCAATGATGTCGCGTCGACGTTGGGTAACGCTTCGCAAGAAACCAGGGGGGAGC TCATTGTCGGCGAATGGTCCTGCGCCCTTCCACCAGAAAGCCTTGCATCCGATTCGAATCAAAATCAAGCGCATA AAGATTTCTGCGGCGGGCAGGTTGATACCTATGGAAACAACACTGCTGGTTGGAGCTTCTGGTGTAAGTCTTGTT TTCCTCTCGTAACTCGAAACTGAAACCAACTGAGCTGAATCTTGCCTTTACAGCTTACACGAAAGAAGAATGCGA TAACGATCCCGGGTGGTGCTTCAAGTCAGCGGTCGGCACTGTTTTACCCACGAAGTTCTTCTCGTACCATCATCA ACAACCAGAGTCCATCCAAAACAGCGTTGCTGGATCCAATCATTCCTTCCTGCTTCATGACGCGGAGCTGCCGTT AACCAGGCGTGACGAGTTTACGAGGCTCTCTATTCCCGAGCGCTTCAAAACCATCGCTAGAAGACGCAGGCGAGA TGGAAACACAGATGCAGATGGTAAAGGGTATGACGACGGCAGGGCGACGGCTGGCATATTTGCTGCGCATAACAT GTCAAGGCTGGGATTCGTCAACCAATACATCTCGGATACCATCGCCAACCTTGGATCCAATGTTATACCCCAGGA ATCTGAAGGTGCTTATACAGAGGGATTCCTTCGTGGTCTTCGGGAGGCAGAGTCGAATTTCTCGACACAAGCTTT CACATCGTAGTGCTTTACCAACCTTGCATTCGCTATTCGATTTTCACCGTTGTATTGCTAGCACCATTTATTTTA CTCTGGGAGTACTTTGCTCGCCCGGTGTTCACAACCAGACATTTCAGTA |
| Length | 2299 |