CopciAB_61145
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_61145 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | eglB | Synonyms | 61145 |
| Uniprot id | Functional description | Belongs to the glycosyl hydrolase 5 (cellulase A) family | |
| Location | scaffold_3:1761009..1763142 | Strand | - |
| Gene length (nt) | 2134 | Transcript length (nt) | 1479 |
| CDS length (nt) | 1203 | Protein length (aa) | 400 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN1285_eglA |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB26262 | 65.8 | 5.776E-182 | 567 |
| Agrocybe aegerita | Agrae_CAA7267109 | 63.6 | 8.001E-177 | 552 |
| Auricularia subglabra | Aurde3_1_1163753 | 60.8 | 7.266E-165 | 518 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1067505 | 60.9 | 3.845E-165 | 518 |
| Pleurotus ostreatus PC9 | PleosPC9_1_116228 | 60.9 | 3.763E-165 | 518 |
| Pleurotus eryngii ATCC 90797 | Pleery1_682179 | 60.9 | 2.429E-163 | 513 |
| Grifola frondosa | Grifr_OBZ72550 | 58.6 | 4.903E-163 | 512 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_124054 | 59.4 | 5.358E-162 | 509 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_191420 | 60.1 | 1.141E-159 | 502 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_85704 | 59.7 | 2.638E-159 | 501 |
| Lentinula edodes NBRC 111202 | Lenedo1_1059630 | 56.1 | 1.173E-153 | 485 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_11376 | 56.1 | 1.164E-153 | 485 |
| Lentinula edodes B17 | Lened_B_1_1_6666 | 58.5 | 2.606E-141 | 449 |
| Schizophyllum commune H4-8 | Schco3_2502504 | 61.4 | 9.023E-140 | 445 |
| Flammulina velutipes | Flave_chr09AA01045 | 57 | 1.911E-118 | 383 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | eglB |
|---|---|
| Protein id | CopciAB_61145.T0 |
| Description | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00734 | Fungal cellulose binding domain | IPR000254 | 26 | 53 |
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 99 | 369 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 22 | 0.9985 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR017853 | Glycoside hydrolase superfamily |
| IPR001547 | Glycoside hydrolase, family 5 |
| IPR035971 | Cellulose-binding domain superfamily |
| IPR000254 | Cellulose-binding domain, fungal |
| IPR018087 | Glycoside hydrolase, family 5, conserved site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
| GO:0005576 | extracellular region | CC |
| GO:0005975 | carbohydrate metabolic process | BP |
| GO:0030248 | cellulose binding | MF |
KEGG
| KEGG Orthology |
|---|
| K01179 |
EggNOG
| COG category | Description |
|---|---|
| G | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| CBM | CBM1 | |
| GH | GH5 | GH5_5 |
Transcription factor
| Group |
|---|
| No records |
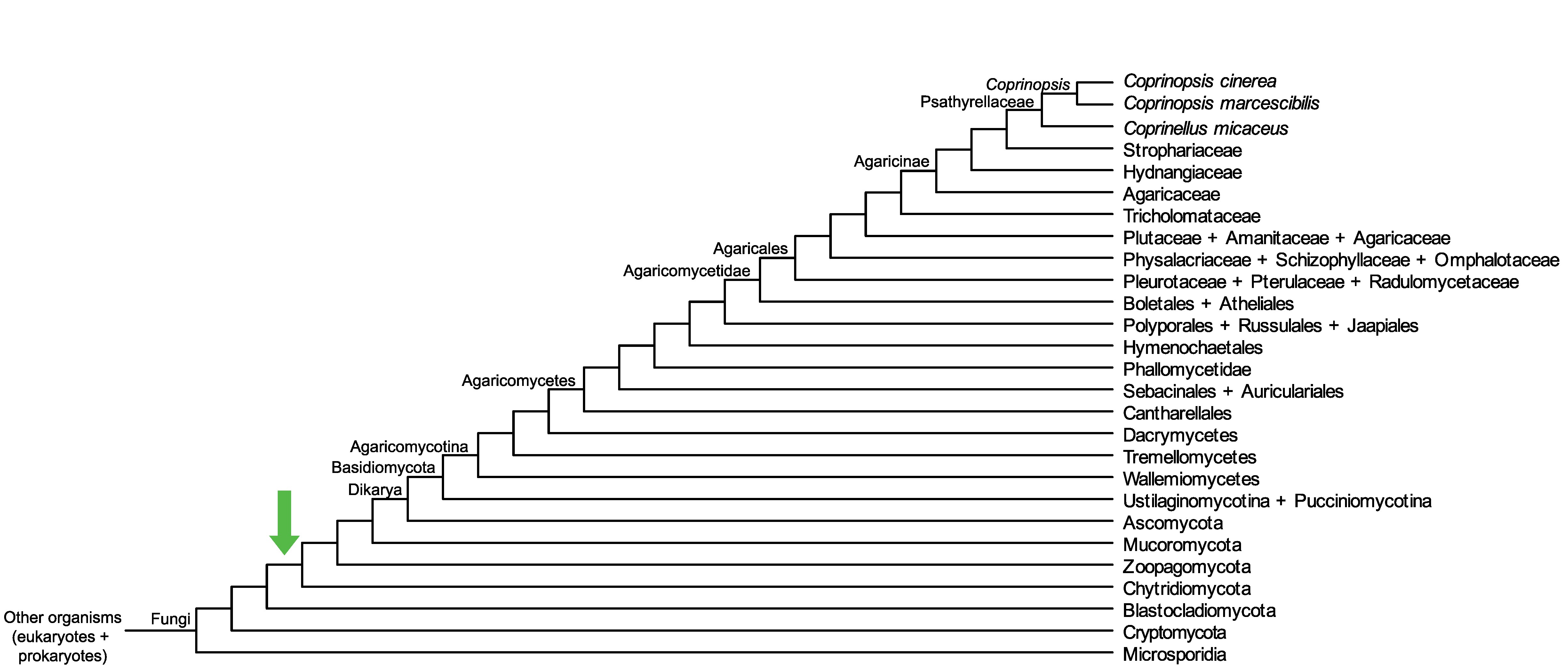
Conservation of CopciAB_61145 across fungi.
Arrow shows the origin of gene family containing CopciAB_61145.
Protein
| Sequence id | CopciAB_61145.T0 |
|---|---|
| Sequence |
>CopciAB_61145.T0 MKSVLSPLSATVIALLSTAAVAVPPYGQCGGSGYTGPSDCDSGYTCVPVNQWYHQCQPGSSAPTNPPNPPVATTT TTAPRPTNSACPGAVTKFRYFGVNQAGAEFGSNIIPGELGTHYIWPAPSSIDFFVNEGFNTFRIPFQLERLNPPA RGLTGSFEQPYLQDLKRTVDYITRNKNSFAIIEPHNYMRYANQTITNVSQFSTWWRNLATEFRDNDKVIFDVMNE PHTMDARVVFDLNQAAVNAIREAGATRQLILVEGTSWTGAWTWTTTSGNGNVFQNIRDPNNNVAIQMHQYLDSDG SGTSPTCVSSSIGRERLAAATAWLKQHNLKGFLGELGAGSNPTCIEAIKGALCSLQESDVWIGWAWWAAGPWWGN YFQSIEPPNGPALAQMYPQAIKPFI |
| Length | 400 |
Coding
| Sequence id | CopciAB_61145.T0 |
|---|---|
| Sequence |
>CopciAB_61145.T0 ATGAAGTCTGTGCTCTCCCCGCTGTCTGCAACTGTGATTGCTCTGTTGAGCACTGCGGCTGTTGCCGTTCCCCCC TACGGCCAGTGCGGTGGCTCTGGATACACTGGACCTTCCGACTGTGACTCTGGCTATACCTGTGTCCCGGTGAAC CAATGGTACCACCAGTGCCAACCCGGTAGCTCCGCCCCCACCAACCCTCCCAATCCCCCTGTCGCTACTACCACT ACCACTGCGCCCAGGCCCACCAACAGTGCTTGCCCAGGCGCCGTCACCAAGTTCCGCTACTTCGGTGTCAACCAA GCTGGTGCTGAGTTCGGCAGCAACATCATCCCCGGAGAGCTCGGGACCCACTACATCTGGCCCGCCCCTAGTTCC ATCGACTTCTTCGTTAACGAAGGTTTCAACACTTTCCGAATTCCCTTCCAATTGGAGCGACTGAACCCCCCTGCT CGTGGGTTGACCGGTTCCTTCGAGCAGCCCTACCTCCAGGACTTGAAGAGGACTGTCGACTACATCACCCGAAAC AAGAACTCTTTCGCCATCATCGAACCCCACAACTACATGCGATATGCCAACCAGACCATCACCAACGTTTCTCAA TTCAGCACCTGGTGGAGGAACCTTGCAACTGAATTCAGGGATAATGACAAGGTTATCTTTGATGTCATGAACGAG CCCCACACTATGGATGCTCGTGTCGTTTTCGACCTCAACCAAGCGGCGGTCAATGCTATCCGTGAAGCTGGTGCT ACCCGCCAATTGATCCTCGTTGAGGGAACCTCCTGGACCGGTGCCTGGACCTGGACTACCACCAGCGGAAACGGC AACGTCTTCCAGAATATCCGTGATCCCAACAACAATGTCGCCATTCAGATGCACCAGTACCTCGACAGTGACGGT TCGGGTACCTCTCCCACTTGTGTTTCCAGCTCCATCGGTCGCGAGCGTCTCGCTGCTGCCACTGCCTGGTTGAAG CAACACAACCTCAAGGGCTTCCTCGGTGAACTCGGTGCTGGTTCCAACCCCACTTGCATTGAGGCCATCAAGGGT GCTCTCTGCTCCCTCCAGGAGTCTGATGTCTGGATTGGTTGGGCCTGGTGGGCTGCTGGTCCCTGGTGGGGCAAC TACTTCCAATCCATTGAGCCCCCCAATGGACCCGCTCTCGCTCAGATGTACCCTCAGGCCATCAAACCCTTCATC TAA |
| Length | 1203 |
Transcript
| Sequence id | CopciAB_61145.T0 |
|---|---|
| Sequence |
>CopciAB_61145.T0 GAACCCATCTCAACCTATCCCGCGCTGTGTTTAACGACTTTATTCTTTCCCGATTCTGCCCATCCATGCGCCCGC GGTGACTTTTTGTCGGCTAAATGACGGATGATAATTTTCTTGGATTTCGTTTATAACCGCGGCGTGGTTTGGTTC CTGAGCTGGCTAGGACGCGTTTCTCGTTCAGGATGAAGTCTGTGCTCTCCCCGCTGTCTGCAACTGTGATTGCTC TGTTGAGCACTGCGGCTGTTGCCGTTCCCCCCTACGGCCAGTGCGGTGGCTCTGGATACACTGGACCTTCCGACT GTGACTCTGGCTATACCTGTGTCCCGGTGAACCAATGGTACCACCAGTGCCAACCCGGTAGCTCCGCCCCCACCA ACCCTCCCAATCCCCCTGTCGCTACTACCACTACCACTGCGCCCAGGCCCACCAACAGTGCTTGCCCAGGCGCCG TCACCAAGTTCCGCTACTTCGGTGTCAACCAAGCTGGTGCTGAGTTCGGCAGCAACATCATCCCCGGAGAGCTCG GGACCCACTACATCTGGCCCGCCCCTAGTTCCATCGACTTCTTCGTTAACGAAGGTTTCAACACTTTCCGAATTC CCTTCCAATTGGAGCGACTGAACCCCCCTGCTCGTGGGTTGACCGGTTCCTTCGAGCAGCCCTACCTCCAGGACT TGAAGAGGACTGTCGACTACATCACCCGAAACAAGAACTCTTTCGCCATCATCGAACCCCACAACTACATGCGAT ATGCCAACCAGACCATCACCAACGTTTCTCAATTCAGCACCTGGTGGAGGAACCTTGCAACTGAATTCAGGGATA ATGACAAGGTTATCTTTGATGTCATGAACGAGCCCCACACTATGGATGCTCGTGTCGTTTTCGACCTCAACCAAG CGGCGGTCAATGCTATCCGTGAAGCTGGTGCTACCCGCCAATTGATCCTCGTTGAGGGAACCTCCTGGACCGGTG CCTGGACCTGGACTACCACCAGCGGAAACGGCAACGTCTTCCAGAATATCCGTGATCCCAACAACAATGTCGCCA TTCAGATGCACCAGTACCTCGACAGTGACGGTTCGGGTACCTCTCCCACTTGTGTTTCCAGCTCCATCGGTCGCG AGCGTCTCGCTGCTGCCACTGCCTGGTTGAAGCAACACAACCTCAAGGGCTTCCTCGGTGAACTCGGTGCTGGTT CCAACCCCACTTGCATTGAGGCCATCAAGGGTGCTCTCTGCTCCCTCCAGGAGTCTGATGTCTGGATTGGTTGGG CCTGGTGGGCTGCTGGTCCCTGGTGGGGCAACTACTTCCAATCCATTGAGCCCCCCAATGGACCCGCTCTCGCTC AGATGTACCCTCAGGCCATCAAACCCTTCATCTAAAGTGCTGAAGGAAGGGAGAAAAGGAGCTCGAGCTTCTGCA AGGATGTAGGATATGTCGTCATTGCTTTGAAATAAACTATGACTTTGAATCTTC |
| Length | 1479 |
Gene
| Sequence id | CopciAB_61145.T0 |
|---|---|
| Sequence |
>CopciAB_61145.T0 GAACCCATCTCAACCTATCCCGCGCTGTGTTTAACGACTTTATTCTTTCCCGATTCTGCCCATCCATGCGCCCGC GGTGACTTTTTGTCGGCTAAATGACGGATGATAATTTTCTTGGATTTCGTTTATAACCGCGGCGTGGTTTGGTTC CTGAGCTGGCTAGGACGCGTTTCTCGTTCAGGATGAAGTCTGTGCTCTCCCCGCTGTCTGCAACTGTGATTGCTC TGTTGAGCACTGCGGCTGTTGCCGTTCCCCCCTACGGCCAGTGCGGTGTAAGTGTCGCACTCAATTGACGGTGGT GTGTTCTCTGACGCTTGCTTCTCTTAGGGCTCTGGATACACTGGACCTTCCGACTGTGACTCTGGCTATACCTGT GTCCCGGTGAACCAATGTACGGCACCTACTCCTACGGCCATTGTTCTCACAAGACTGACTGCCGTTTGAATTAGG GTACCACCAGTGCCAACCCGGTAGCTCCGCCCCCACCAACCCTCCCAATCCCCCTGTCGCTACTACCACTACCAC TGCGCCCAGGCCCACCAACAGTGCTTGCCCAGGCGCCGTCACCAAGTTCCGCTACTTCGGTGTCAACCAAGCTGG TGCTGAGTTCGGCAGCAACATCATCCCCGTACGCTGCTCTCCTTCCACCATCCCCTGACAGTTGAACTTACGCCC GACGATGCTCGGTTTCAGGGAGAGCTCGGGACCCACTACATCTGGCCCGCCCCTAGTTCCATCGACTTCTTCGTT AACGAAGGTTTCAACACTTTCCGAATTCCCTTCCAATTGGAGCGACTGAACCCCCCTGCTCGTGGGTTGACCGGT TCCTTCGAGCAGCCCTACCTCCAGGACTTGAAGAGGACTGTCGACTACATCACCCGAAACAAGAACTCTTTCGCC ATCATCGAACGTGTGTTATTTACTCTTTCTGACACTTGGTTTGTGGAGACTGAAGGGGACATGCAGCCCACAACT ACATGCGATATGCCAACCAGACCATCACCAACGTTTCTCAGTGAGTTATTTACCTCTGAGAGGATATAAACTGTT GCTTACGCTTTGTCGTTATAGATTCAGCACCTGTAAGTTTCTCGCATCGCTTGAGACCTGTTCCAGAGATTTAAC CGTGTTGCGTAGGGTGGAGGAACCTTGCAACTGAATTCGTAAGTTCGATTTTTCCATTGGAAACAAGTTCACCCT CTGACTTTGTTTCTAGAGGGATAATGACAAGGTTATCTTTGGTGTGTATCCATCGAAGCCAAGCGGGCAAGAAAG AGATCTAAACACTGTCTAATAGATGTCATGAACGAGCCCCACACTATGGATGCTCGTGTCGTTTTCGACCTCAAC CAAGCGGCGGTCAATGCTATCCGTGAAGCTGGTGCTACCCGCCAATTGATCCTCGTTGAGGGAACCTCCTGGACC GGTGCCTGGAGTAAGCGATCCACCTTTTCCTCGATAGAGAATTACCTCATTGACATGTTGACGGAATTTAGCCTG GACTACCACCAGCGGAAACGGCAACGTCTTCCAGAATATCCGTGATCCCAACAACAATGTCGCCATTCGTACGTC TTGTTCATCGACATCTCCTTCTAACAATTCCTGATCAACGTTTTGGCACCACTCGTTACCACTTTCAGAGATGCA CCAGTACCTCGACAGTGACGGTTCGGGTACCTCTCCCACTTGTGTTTCCAGCTCCATCGGTCGCGAGCGTCTCGC TGCTGCCACTGCCTGGTTGAAGCAACACAACCTCAAGGGCTTCCTCGGTGAACTCGGTGCTGGTTCCAACCCCAC TTGCATTGAGGCCATCAAGGGTGCTCTCTGCTCCCTCCAGGAGTCTGATGTCTGGATTGGTTGGGCCTGGTGGGC TGCTGGTCCCTGGTGGGGCAACGTAAGTTGTTTCTGCCTTTCGTGTGGACGGGATGCTGATGCAATGTTTTATTT TCTTCCTTATAGTACTTCCAATCCATTGAGCCCCCCAATGGACCCGCTCTCGCTCAGATGTACCCTCAGGCCATC AAACCCTTCATCTAAAGTGCTGAAGGAAGGGAGAAAAGGAGCTCGAGCTTCTGCAAGGATGTAGGATATGTCGTC ATTGCTTTGAAATAAACTATGACTTTGAATCTTC |
| Length | 2134 |