CopciAB_71850
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_71850 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 71850 |
| Uniprot id | Functional description | Domain in the RNA-binding Lupus La protein; unknown function | |
| Location | scaffold_10:430663..433865 | Strand | + |
| Gene length (nt) | 3203 | Transcript length (nt) | 1972 |
| CDS length (nt) | 1569 | Protein length (aa) | 522 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Schizosaccharomyces pombe | sla1 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7261977 | 58.1 | 3.124E-184 | 580 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB17657 | 56 | 1.726E-183 | 578 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_195118 | 57.2 | 5.326E-183 | 576 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_116412 | 57.2 | 2.723E-182 | 574 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1059064 | 54.4 | 1.04E-166 | 529 |
| Pleurotus ostreatus PC9 | PleosPC9_1_113124 | 54.4 | 1.018E-166 | 529 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1484229 | 56.8 | 8.274E-162 | 515 |
| Lentinula edodes NBRC 111202 | Lenedo1_1061492 | 46.3 | 2.484E-147 | 473 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_21619 | 46.4 | 2.465E-147 | 473 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_120098 | 50.5 | 5.884E-145 | 466 |
| Schizophyllum commune H4-8 | Schco3_2639628 | 46.6 | 2.246E-129 | 421 |
| Flammulina velutipes | Flave_chr11AA01297 | 46.6 | 2.439E-125 | 409 |
| Auricularia subglabra | Aurde3_1_1279080 | 40 | 3.867E-103 | 345 |
| Grifola frondosa | Grifr_OBZ74753 | 44.1 | 5.779E-100 | 335 |
| Lentinula edodes B17 | Lened_B_1_1_15359 | 45.9 | 1.465E-94 | 319 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_71850.T0 |
| Description | Domain in the RNA-binding Lupus La protein; unknown function |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd08029 | LA_like_fungal | - | 58 | 143 |
| CDD | cd12291 | RRM1_La | - | 157 | 231 |
| Pfam | PF05383 | La domain | IPR006630 | 62 | 119 |
| Pfam | PF00076 | RNA recognition motif. (a.k.a. RRM, RBD, or RNP domain) | IPR000504 | 171 | 228 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR014886 | La protein, xRRM domain |
| IPR036390 | Winged helix DNA-binding domain superfamily |
| IPR012677 | Nucleotide-binding alpha-beta plait domain superfamily |
| IPR002344 | Lupus La protein |
| IPR035979 | RNA-binding domain superfamily |
| IPR000504 | RNA recognition motif domain |
| IPR045180 | La domain containing protein |
| IPR036388 | Winged helix-like DNA-binding domain superfamily |
| IPR006630 | La-type HTH domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0003723 | RNA binding | MF |
| GO:0005634 | nucleus | CC |
| GO:0006396 | RNA processing | BP |
| GO:1990904 | ribonucleoprotein complex | CC |
| GO:0003676 | nucleic acid binding | MF |
KEGG
| KEGG Orthology |
|---|
| K11090 |
EggNOG
| COG category | Description |
|---|---|
| A | Domain in the RNA-binding Lupus La protein; unknown function |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| winged helix-turn-helix |
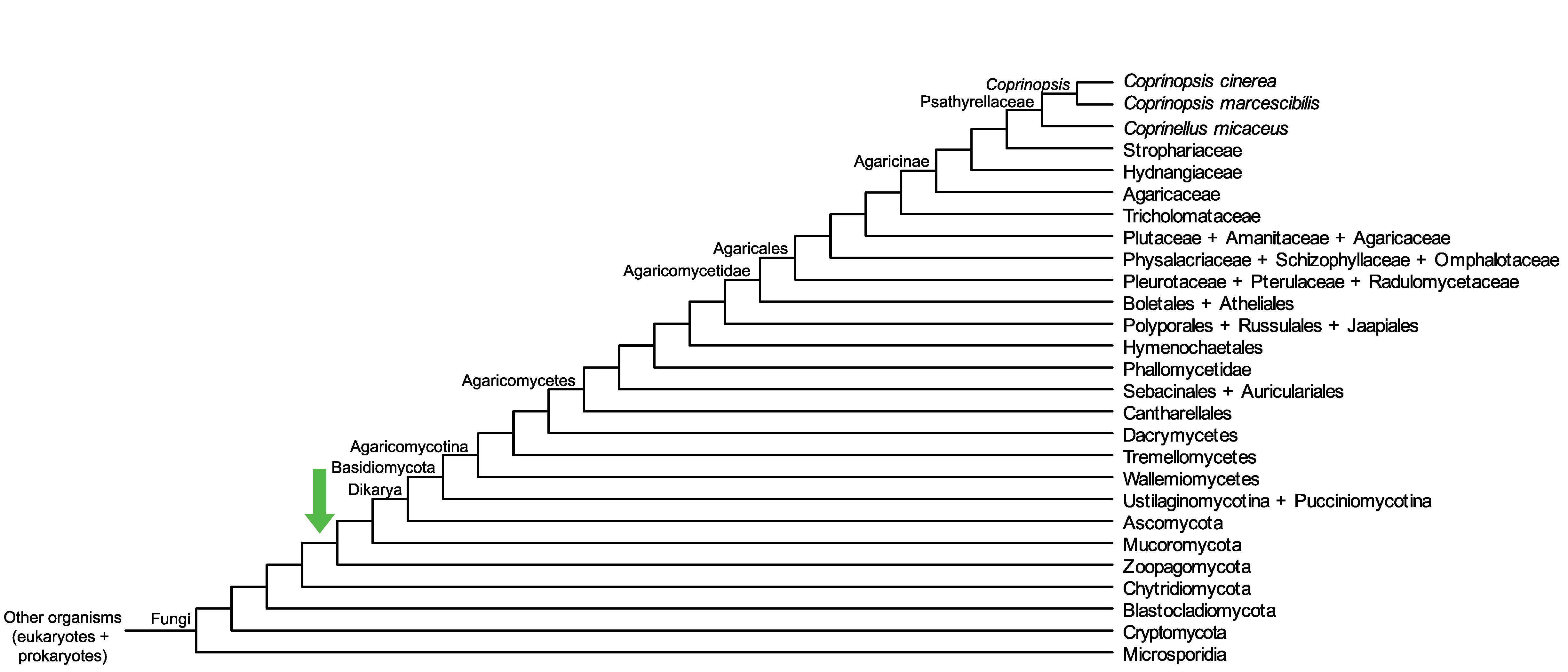
Conservation of CopciAB_71850 across fungi.
Arrow shows the origin of gene family containing CopciAB_71850.
Protein
| Sequence id | CopciAB_71850.T0 |
|---|---|
| Sequence |
>CopciAB_71850.T0 MSGEDTSATTMAVDAAPTTETPASTTTTTTAAAPSETTTTTTTNPEKPIEDGEALAKMEKARQQVEFYFADSNLP YDKFMWTLHTKDPEHWIPLSTIASFKRMREYTALGAEFVLNALRPSTFLELDESGREGAELAGVKVRRKTEVREV KDAFERSVYAKGFPEEDPTLQNRLEEFFATFGKVNSVRMRRDEHKKFKTSVFAEFADFESVDKFLKADPKPTFEG KELLIMSKHDYCEMKIKEKGLTGKAANHRREMLTKGRGFDAFRESSTSSPSSSSKDEPKEILLEYIGNRIPIKKD NEGNGYVEESSIPFVRGATLRFDGCAGDCTWSDIKDPIKTLFDGKPPYIKFTRGDDFGLVGFHKALSEEEVEKVK EAVRRVNGREVRWSVPGEEEERVFQVERAQVAARGAMAQITGGGKKAAGGGRGGRGGGRGGRGGRGGRGGGRGGG RDNGRKENGKEASGGAKEEKGRAEGGDVEVGEKRKRAVEPDGGPYTGVRGTNAPPAVVASSESAAKKAKTDA |
| Length | 522 |
Coding
| Sequence id | CopciAB_71850.T0 |
|---|---|
| Sequence |
>CopciAB_71850.T0 ATGTCTGGAGAAGATACTTCTGCTACTACTATGGCCGTCGACGCGGCTCCTACGACGGAGACTCCCGCCTCTACC ACCACCACTACGACCGCAGCTGCTCCCTCAGAAACGACTACGACCACCACGACCAACCCCGAAAAACCCATCGAA GACGGGGAAGCGCTCGCCAAGATGGAAAAAGCTCGTCAACAAGTCGAGTTCTACTTTGCAGATTCGAATTTGCCT TATGATAAATTCATGTGGACCCTCCACACCAAAGACCCCGAACACTGGATCCCCCTCTCCACCATCGCCTCCTTC AAACGCATGCGCGAGTACACCGCCCTCGGCGCCGAGTTCGTCTTGAACGCCCTGCGCCCCTCGACCTTCCTCGAG CTCGATGAGAGTGGGAGGGAGGGTGCCGAGTTGGCTGGTGTGAAGGTTAGGAGGAAGACGGAGGTTAGGGAGGTG AAGGATGCGTTTGAGAGGAGTGTTTATGCTAAAGGGTTCCCGGAGGAAGACCCCACACTGCAGAACCGCCTCGAA GAGTTCTTTGCGACGTTCGGCAAGGTGAATTCGGTTAGGATGAGGAGGGACGAGCATAAAAAGTTCAAGACCTCC GTATTCGCAGAATTCGCCGACTTTGAGTCTGTGGACAAGTTCCTCAAGGCTGATCCCAAACCAACGTTTGAAGGG AAGGAGTTGTTGATTATGTCCAAACACGACTACTGCGAAATGAAAATCAAAGAAAAAGGTCTAACCGGAAAGGCC GCCAACCACCGCCGCGAAATGCTCACCAAAGGACGGGGATTCGACGCCTTCCGCGAGTCCTCTACCTCCTCCCCT TCTTCCTCCTCCAAAGACGAACCCAAAGAAATCCTCCTCGAATACATCGGAAACCGTATCCCGATCAAGAAAGAT AATGAAGGGAACGGCTATGTGGAAGAATCCTCCATCCCTTTCGTACGTGGTGCGACACTCCGATTCGACGGCTGC GCCGGGGACTGTACGTGGTCCGACATCAAAGACCCTATCAAAACGCTCTTCGATGGTAAACCACCCTATATCAAG TTTACTCGAGGTGATGATTTTGGGTTGGTGGGATTCCATAAGGCGTTGAGTGAGGAGGAGGTGGAGAAGGTTAAG GAGGCGGTGAGGAGGGTTAATGGGAGGGAGGTGAGGTGGAGTGTGCCCGGGGAGGAGGAGGAGAGGGTGTTCCAG GTTGAGAGGGCGCAGGTCGCGGCGAGGGGGGCGATGGCGCAGATTACGGGAGGGGGGAAGAAGGCTGCAGGTGGT GGACGCGGTGGCCGGGGAGGTGGACGTGGTGGACGAGGAGGAAGGGGAGGTCGGGGTGGAGGACGAGGCGGAGGA CGGGATAACGGGAGGAAGGAGAATGGCAAGGAGGCTTCAGGCGGGGCGAAGGAGGAGAAGGGCAGAGCTGAGGGA GGAGATGTAGAGGTTGGTGAGAAGAGGAAGAGGGCTGTTGAGCCTGATGGGGGACCGTATACGGGTGTGAGGGGT ACGAATGCCCCGCCTGCGGTTGTTGCGTCTAGTGAGAGTGCGGCGAAGAAGGCCAAGACGGATGCTTGA |
| Length | 1569 |
Transcript
| Sequence id | CopciAB_71850.T0 |
|---|---|
| Sequence |
>CopciAB_71850.T0 AACCTCCTCCTTCTCTCTCAAAAATTCTGCTAGGACGCTTACGCTTATCCTGCGATAAGCCTCACTCTCTAAAGG GATATTATCCGACTGGAGCGATAGGAAGATCCACTTTACCAGTTCTTAGGACGATATAGGGAAGAGGTTTGAAAA ACTTGTTGAAGAAGAACTTGAAGTGAAGAAATGTCTGGAGAAGATACTTCTGCTACTACTATGGCCGTCGACGCG GCTCCTACGACGGAGACTCCCGCCTCTACCACCACCACTACGACCGCAGCTGCTCCCTCAGAAACGACTACGACC ACCACGACCAACCCCGAAAAACCCATCGAAGACGGGGAAGCGCTCGCCAAGATGGAAAAAGCTCGTCAACAAGTC GAGTTCTACTTTGCAGATTCGAATTTGCCTTATGATAAATTCATGTGGACCCTCCACACCAAAGACCCCGAACAC TGGATCCCCCTCTCCACCATCGCCTCCTTCAAACGCATGCGCGAGTACACCGCCCTCGGCGCCGAGTTCGTCTTG AACGCCCTGCGCCCCTCGACCTTCCTCGAGCTCGATGAGAGTGGGAGGGAGGGTGCCGAGTTGGCTGGTGTGAAG GTTAGGAGGAAGACGGAGGTTAGGGAGGTGAAGGATGCGTTTGAGAGGAGTGTTTATGCTAAAGGGTTCCCGGAG GAAGACCCCACACTGCAGAACCGCCTCGAAGAGTTCTTTGCGACGTTCGGCAAGGTGAATTCGGTTAGGATGAGG AGGGACGAGCATAAAAAGTTCAAGACCTCCGTATTCGCAGAATTCGCCGACTTTGAGTCTGTGGACAAGTTCCTC AAGGCTGATCCCAAACCAACGTTTGAAGGGAAGGAGTTGTTGATTATGTCCAAACACGACTACTGCGAAATGAAA ATCAAAGAAAAAGGTCTAACCGGAAAGGCCGCCAACCACCGCCGCGAAATGCTCACCAAAGGACGGGGATTCGAC GCCTTCCGCGAGTCCTCTACCTCCTCCCCTTCTTCCTCCTCCAAAGACGAACCCAAAGAAATCCTCCTCGAATAC ATCGGAAACCGTATCCCGATCAAGAAAGATAATGAAGGGAACGGCTATGTGGAAGAATCCTCCATCCCTTTCGTA CGTGGTGCGACACTCCGATTCGACGGCTGCGCCGGGGACTGTACGTGGTCCGACATCAAAGACCCTATCAAAACG CTCTTCGATGGTAAACCACCCTATATCAAGTTTACTCGAGGTGATGATTTTGGGTTGGTGGGATTCCATAAGGCG TTGAGTGAGGAGGAGGTGGAGAAGGTTAAGGAGGCGGTGAGGAGGGTTAATGGGAGGGAGGTGAGGTGGAGTGTG CCCGGGGAGGAGGAGGAGAGGGTGTTCCAGGTTGAGAGGGCGCAGGTCGCGGCGAGGGGGGCGATGGCGCAGATT ACGGGAGGGGGGAAGAAGGCTGCAGGTGGTGGACGCGGTGGCCGGGGAGGTGGACGTGGTGGACGAGGAGGAAGG GGAGGTCGGGGTGGAGGACGAGGCGGAGGACGGGATAACGGGAGGAAGGAGAATGGCAAGGAGGCTTCAGGCGGG GCGAAGGAGGAGAAGGGCAGAGCTGAGGGAGGAGATGTAGAGGTTGGTGAGAAGAGGAAGAGGGCTGTTGAGCCT GATGGGGGACCGTATACGGGTGTGAGGGGTACGAATGCCCCGCCTGCGGTTGTTGCGTCTAGTGAGAGTGCGGCG AAGAAGGCCAAGACGGATGCTTGAGGTGGGCGTACATCAAGGGCGTTGGTCGTCCTTCATCTAGGGTGTTTGGTG GTACCCTTCATCGCTAACCGCACCGCTGGGTTCTCCGCCTCTCAAAAATCGAAATGAAATGTTATCTATGTAGCA CTAGTTTCTGCTTTTTTGTTGTGTTTGTTTCGTCTGTAGTATGTAGTACCACGTAGCTAGGTCGGGACGACTGAA CGAATTGCATATTTATCTTACC |
| Length | 1972 |
Gene
| Sequence id | CopciAB_71850.T0 |
|---|---|
| Sequence |
>CopciAB_71850.T0 AACCTCCTCCTTCTCTCTCAAAAATTCTGCTAGGACGCTTACGCTTATCCTGCGATAAGCCTCACTCTCTAAAGG GATATTATCCGACTGGAGCGATAGGAAGATCCACTTTACCAGTTCTTAGGACGATATAGGGAAGAGGTTTGAAAA ACTTGTTGAAGAAGAACTTGAAGTGAAGAAATGTCTGGAGAAGATACTTCTGCTACTACTATGGCCGTCGACGCG GCTCCTACGACGGAGACTCCCGCCTCTACCACCACCACTACGACCGCAGCTGCTCCCTCAGAAACGACTACGACC ACCACGACCAACCCCGAAAAACCCATCGAAGACGGGGAAGCGCTCGCCAAGATGGAAAAAGCTCGTCAACAAGGT GCGTATTTATCTATCTGTGTGTTGTGAATGGGGAATGACTGATTGTGATTTTGGGAATGGTACAGTCGAGTTCTA CTTTGCAGATTCGAATTTGCCTTATGATAAGTGCGTTCCCTCCCTTCCCTCCCTCCTTCTTCCCCTTTCCCCCTT GCCCCTTCCCCCTCCCTCCTCCTTCCCCCATCCCCTCACAAACCCCCTCCCACTCCAACCCCCACTAACCCCTCC TCTCTCCCTCCTCCAGATTCATGTGGACCCTCCACACCAAAGACCCCGAACACTGGATCCCCCTCTCCACCATCG CCTCCTTCAAACGCATGCGCGAGTACACCGCCCTCGGCGCCGAGTTCGTCTTGAACGCCCTGCGCCCCTCGACCT TCCTCGAGCTCGATGAGAGTGGGAGGGAGGGTGCCGAGTTGGCTGGTGTGAAGGTTAGGAGGAAGACGGAGGTTA GGGAGGTGAAGGATGCGTTTGAGAGGAGTGTTTATGCTGTGAGTTCCTTTGCGTCCCTGTGTGTGTGGAAGTGGA AGTAAGTAGTGTTGATTGATGCGTTTGGTATTTTGTTTTCTTTTTGTAGAAAGGGTTCCCGGAGGAAGACCCCAC ACTGCAGAACCGCCTCGAAGAGTTCTTTGCGACGTTCGGCAAGGTGAATTCGGTTAGGATGAGGAGGGACGAGCA TAAAAAGTTCAAGGTGTGTCTTCCTTCCCGCATGAACCCTTCTGTTTATTTTGTTCCTTTTCTTTATCCCCTTCT GTTTATCCTCCTGTATGGCCTCGTTCGATTCTTGCGTACCGCCTTTACCATGTCAATCGTGTTTTTTTCCCTCCC ATCGCTATTGTGTTCTCGCGCGTTTTCGCTGCCGGTGTCCATTTTGTTGTCCCGCTTCTCCACTTGCCCCTGCTC CCAGCGAACGAGTCTCCAGGATATCCGTTTCCCCATCGACTCGGACGTACCCAGTCGAAACCTGGAGCATCGACA CCGAATTCCCTCGTTTGATTTCCCTGTCACCCTATTCTTCGCTGCTCTGCTTCGTGTTTCGTGAAATCCTCTGAA TCATCTCGTCTTCTTCTTCAATGGCGTTCCCGCCCCACCTCCCTCTCAACGCCCAACGCGTCCCCGTGTTTATTT GTTTGCTGGGTGAGCTGGGGAATGGGTCGTGCAGAACGGGGATGGCCGGTGCCCGCTGTCGCTGGACCAGAAGTT CTGTTGTCCCACCCGGTCGTGACATTTGAGCTCTTCCATCACTTTCGACATACATGCTCTTTTTGCCTGGTGCAC ACAAACGTTCCTTTTCCCTGATTGCACACGACTGTCCTTTTTTCTGGTTACACACGACCGTCCTTTTTCCTGGTT GCACACGACGAACCCATCCGTATATCATTATGCCTAACTGACACTCCCCCTCCCACTTGCCAGACCTCCGTATTC GCAGAATTCGCCGACTTTGAGTCTGTGGACAAGTTCCTCAAGGCTGATCCCAAACCAACGTTTGAAGGGAAGGAG TTGTTGATTATGTCCAAGTGCGTCGTCTCTTCCTCTCGTTGGGATTTTCTCCTTGGCTTTTATGAGCCTGTCTCC TTGCGTTTTCGACCCTCTTCCTCGCAGGATTGCCTCGCTCAGTTGTACCACTTCGCGGCTGGTCATTCGATCGCC TCGTTCCATGAACTCGATATTTGACGCCCCTTTGCCCTTCTCGTTAATGTCTACTTGCTTACTTACACCCTACAC TACCCCCAGACACGACTACTGCGAAATGAAAATCAAAGAAAAAGGTCTAACCGGAAAGGCCGCCAACCACCGCCG CGAAATGCTCACCAAAGGACGGGGATTCGACGCCTTCCGCGAGTCCTCTACCTCCTCCCCTTCTTCCTCCTCCAA AGACGAACCCAAAGAAATCCTCCTCGAATACATCGGAAACCGTATCCCGATCAAGAAAGATAATGAAGGGAACGG CTATGTGGAAGAATCCTCCATCCCTTTCGTACGTGGTGCGACACTCCGATTCGACGGCTGCGCCGGGGACTGTAC GTGGTCCGACATCAAAGACCCTATCAAAACGCTCTTCGATGGTAAACCACCCTATATCAAGTTTACTCGAGGTGA TGATTTTGGGTTGGTGGGATTCCATAAGGCGTTGAGTGAGGAGGAGGTGGAGAAGGTTAAGGAGGCGGTGAGGAG GGTTAATGGGAGGGAGGTGAGGTGGAGTGTGCCCGGGGAGGAGGAGGAGAGGGTGTTCCAGGTTGAGAGGGCGCA GGTCGCGGCGAGGGGGGCGATGGCGCAGATTACGGGAGGGGGGAAGAAGGCTGCAGGTGGTGGACGCGGTGGCCG GGGAGGTGGACGTGGTGGACGAGGAGGAAGGGGAGGTCGGGGTGGAGGACGAGGCGGAGGACGGGATAACGGGAG GAAGGAGAATGGCAAGGAGGCTTCAGGCGGGGCGAAGGAGGAGAAGGGCAGAGCTGAGGGAGGAGATGTAGAGGT TGGTGAGAAGAGGAAGAGGGCTGTTGAGCCTGATGGGGGACCGTATACGGGTGTGAGGGGTACGAATGCCCCGCC TGCGGTTGTTGCGTCTAGTGAGAGTGCGGCGAAGAAGGCCAAGACGGATGCTTGAGGTGGGCGTACATCAAGGGC GTTGGTCGTCCTTCATCTAGGGTGTTTGGTGGTACCCTTCATCGCTAACCGCACCGCTGGGTTCTCCGCCTCTCA AAAATCGAAATGAAATGTTATCTATGTAGCACTAGTTTCTGCTTTTTTGTTGTGTTTGTTTCGTCTGTAGTATGT AGTACCACGTAGCTAGGTCGGGACGACTGAACGAATTGCATATTTATCTTACC |
| Length | 3203 |